The Educational Program of Macrophages toward a Hyperprogressive Disease-Related Phenotype Is Orchestrated by Tumor-Derived Extracellular Vesicles
- PMID: 36555441
- PMCID: PMC9779478
- DOI: 10.3390/ijms232415802
The Educational Program of Macrophages toward a Hyperprogressive Disease-Related Phenotype Is Orchestrated by Tumor-Derived Extracellular Vesicles
Abstract
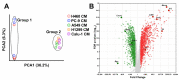
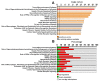
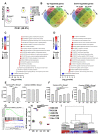
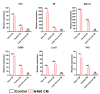
Hyperprogressive disease (HPD), an aggressive acceleration of tumor growth, was observed in a group of cancer patients treated with anti-PD1/PDL1 antibodies. The presence of a peculiar macrophage subset in the tumor microenvironment is reported to be a sort of "immunological prerequisite" for HPD development. These macrophages possess a unique phenotype that it is not clear how they acquire. We hypothesized that certain malignant cells may promote the induction of an "HPD-related" phenotype in macrophages. Bone-marrow-derived macrophages were exposed to the conditioned medium of five non-small cell lung cancer cell lines. Macrophage phenotype was analyzed by microarray gene expression profile and real-time PCR. We found that human NSCLC cell lines, reported as undergoing HPD-like tumor growth in immunodeficient mice, polarized macrophages towards a peculiar pro-inflammatory phenotype sharing both M1 and M2 features. Lipid-based factors contained in cancer cell-conditioned medium induced the over-expression of several pro-inflammatory cytokines and the activation of innate immune receptor signaling pathways. We also determined that tumor-derived Extracellular Vesicles represent the main components involved in the observed macrophage re-education program. The present study might represent the starting point for the future development of diagnostic tools to identify potential hyperprogressors.
Keywords: anti-PD1 antibody; extracellular vesicles; hyperprogressive disease; immune checkpoint inhibitors; macrophages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery.J Exp Clin Cancer Res. 2019 Feb 8;38(1):62. doi: 10.1186/s13046-019-1027-0. J Exp Clin Cancer Res. 2019. PMID: 30736829 Free PMC article.
-
Effect of colorectal cancer-derived extracellular vesicles on the immunophenotype and cytokine secretion profile of monocytes and macrophages.Cell Commun Signal. 2018 Apr 24;16(1):17. doi: 10.1186/s12964-018-0229-y. Cell Commun Signal. 2018. PMID: 29690889 Free PMC article.
-
Tumor Cells Modulate Macrophage Phenotype in a Novel In Vitro Co-Culture Model of the NSCLC Tumor Microenvironment.J Thorac Oncol. 2022 Oct;17(10):1178-1191. doi: 10.1016/j.jtho.2022.06.011. Epub 2022 Jul 5. J Thorac Oncol. 2022. PMID: 35798240 Free PMC article.
-
Managing Hyperprogressive Disease in the Era of Programmed Cell Death Protein 1/Programmed Death-Ligand 1 Blockade: A Case Discussion and Review of the Literature.Oncologist. 2020 May;25(5):369-374. doi: 10.1634/theoncologist.2019-0671. Epub 2020 Feb 24. Oncologist. 2020. PMID: 32091646 Free PMC article. Review.
-
Hyperprogressive Disease upon Immune Checkpoint Blockade: Focus on Non-small Cell Lung Cancer.Curr Oncol Rep. 2020 Apr 16;22(5):41. doi: 10.1007/s11912-020-00908-9. Curr Oncol Rep. 2020. PMID: 32296957 Review.
Cited by
-
Impact of in vitro SARS-CoV-2 infection on breast cancer cells.Sci Rep. 2024 Jun 7;14(1):13134. doi: 10.1038/s41598-024-63804-3. Sci Rep. 2024. PMID: 38849411 Free PMC article.
References
-
- Champiat S., Dercle L., Ammari S., Massard C., Hollebecque A., Postel-Vinay S., Chaput N., Eggermont A., Marabelle A., Soria J.-C., et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clin. Cancer Res. 2017;23:1920–1928. doi: 10.1158/1078-0432.CCR-16-1741. - DOI - PubMed
-
- Camelliti S., Le Noci V., Bianchi F., Moscheni C., Arnaboldi F., Gagliano N., Balsari A., Garassino M.C., Tagliabue E., Sfondrini L., et al. Mechanisms of hyperprogressive disease after immune checkpoint inhibitor therapy: What we (don’t) know. J. Exp. Clin. Cancer Res. 2020;39:236. doi: 10.1186/s13046-020-01721-9. - DOI - PMC - PubMed
-
- Lo Russo G., Moro M., Sommariva M., Cancila V., Boeri M., Centonze G., Ferro S., Ganzinelli M., Gasparini P., Huber V., et al. Antibody-Fc/FcR Interaction on Macrophages as a Mechanism for Hyperprogressive Disease in Non-small Cell Lung Cancer Subsequent to PD-1/PD-L1 Blockade. Clin. Cancer Res. 2019;25:989–999. doi: 10.1158/1078-0432.CCR-18-1390. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

