In-Silico Functional Metabolic Pathways Associated to Chlamydia trachomatis Genital Infection
- PMID: 36555488
- PMCID: PMC9781786
- DOI: 10.3390/ijms232415847
In-Silico Functional Metabolic Pathways Associated to Chlamydia trachomatis Genital Infection
Abstract
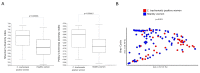
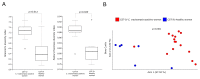
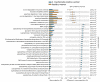
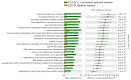
The advent of high-throughput technologies, such as 16s rDNA sequencing, has significantly contributed to expanding our knowledge of the microbiota composition of the genital tract during infections such as Chlamydia trachomatis. The growing body of metagenomic data can be further exploited to provide a functional characterization of microbial communities via several powerful computational approaches. Therefore, in this study, we investigated the predicted metabolic pathways of the cervicovaginal microbiota associated with C. trachomatis genital infection in relation to the different Community State Types (CSTs), via PICRUSt2 analysis. Our results showed a more rich and diverse mix of predicted metabolic pathways in women with a CST-IV microbiota as compared to all the other CSTs, independently from infection status. C. trachomatis genital infection further modified the metabolic profiles in women with a CST-IV microbiota and was characterized by increased prevalence of the pathways for the biosynthesis of precursor metabolites and energy, biogenic amino-acids, nucleotides, and tetrahydrofolate. Overall, predicted metabolic pathways might represent the starting point for more precisely designed future metabolomic studies, aiming to investigate the actual metabolic pathways characterizing C. trachomatis genital infection in the cervicovaginal microenvironment.
Keywords: Chlamydia trachomatis; PICRUSt2; cervicovaginal microbiota; in-silico metabolic profiling; metagenomic data.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Vaginal microbiota composition and association with prevalent Chlamydia trachomatis infection: a cross-sectional study of young women attending a STI clinic in France.Sex Transm Infect. 2018 Dec;94(8):616-618. doi: 10.1136/sextrans-2017-053346. Epub 2018 Jan 22. Sex Transm Infect. 2018. PMID: 29358524
-
Effects of azithromycin and doxycycline on the vaginal microbiota of women with urogenital Chlamydia trachomatis infection: a substudy of the Chlazidoxy randomized controlled trial.Clin Microbiol Infect. 2023 Aug;29(8):1056-1062. doi: 10.1016/j.cmi.2023.04.020. Epub 2023 Apr 24. Clin Microbiol Infect. 2023. PMID: 37100225 Clinical Trial.
-
The Cervicovaginal Microbiota in Women Notified for Chlamydia trachomatis Infection: A Case-Control Study at the Sexually Transmitted Infection Outpatient Clinic in Amsterdam, The Netherlands.Clin Infect Dis. 2017 Jan 1;64(1):24-31. doi: 10.1093/cid/ciw586. Epub 2016 Aug 27. Clin Infect Dis. 2017. PMID: 27567124
-
Cervicovaginal microbiota in Chlamydia trachomatis and other preventable sexually transmitted infections of public health importance: a systematic umbrella review.New Microbiol. 2025 May;48(1):5-13. New Microbiol. 2025. PMID: 40314676
-
Chlamydia trachomatis Genital Tract Infections: When Host Immune Response and the Microbiome Collide.Trends Microbiol. 2016 Sep;24(9):750-765. doi: 10.1016/j.tim.2016.05.007. Epub 2016 Jun 16. Trends Microbiol. 2016. PMID: 27320172 Review.
Cited by
-
Metagenomic analysis reveals the novel role of vaginal Lactobacillus iners in Chinese healthy pregnant women.NPJ Biofilms Microbiomes. 2025 May 30;11(1):92. doi: 10.1038/s41522-025-00731-9. NPJ Biofilms Microbiomes. 2025. PMID: 40447596 Free PMC article.
References
-
- Filardo S., Scalese G., Virili C., Pontone S., di Pietro M., Covelli A., Bedetti G., Marinelli P., Bruno G., Stramazzo I., et al. The Potential Role of Hypochlorhydria in the Development of Duodenal Dysbiosis: A Preliminary Report. Front. Cell. Infect. Microbiol. 2022;12:854904. doi: 10.3389/fcimb.2022.854904. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

