A Pan-Cancer Landscape of ABCG2 across Human Cancers: Friend or Foe?
- PMID: 36555598
- PMCID: PMC9784838
- DOI: 10.3390/ijms232415955
A Pan-Cancer Landscape of ABCG2 across Human Cancers: Friend or Foe?
Abstract
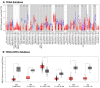
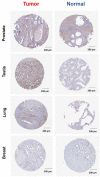
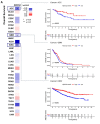
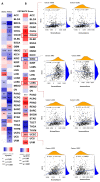
Emerging evidence from research or clinical studies reported that ABCG2 (ATP-binding cassette sub-family G member 2) interrelates with multidrug resistance (MDR) development in cancers. However, no comprehensive pan-cancer analysis is available at present. Therefore, we explore multiple databases, such as TCGA to investigate the potential therapeutic roles of ABCG2 across 33 different tumors. ABCG2 is expressed on a lower level in most cancers and shows a protective effect. For example, a lower expression level of ABCG2 was detrimental to the survival of adrenocortical carcinoma (TCGA-ACC), glioblastoma multiforme (GBM), and kidney renal clear cell carcinoma (KIRC) patients. Distinct associations exist between ABCG2 expression and stemness scores, microenvironmental scores, microsatellite instability (MSI), and tumor mutational burden (TMB) of tumor patients. We observed a significant positive correlation between the ABCG2 mutation site and prognosis in uterine corpus endometrial carcinoma (UCEC) patients. Moreover, transmembrane transporter activity and hormone biosynthetic-associated functions were found to be involved in the functionality of ABCG2 and its related genes. The cDNAs of cancer cell lines were collected to detect exon mutation sequences and to analyze ABCG2 mRNA expression. The mRNA expression level of ABCG2 showed a significant difference among spheres and drug-resistant cancer cell lines compared with their corresponding adherent cancer cell lines in six types of cancer. This pan-cancer study provides, for the first time, a comprehensive understanding of the multifunctionality of ABCG2 and unveils further details of the potential therapeutic role of ABCG2 in pan-cancer.
Keywords: ABCG2; cancer stem cell; genetic alteration; pan-cancer; prognosis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflict of interest.
Figures










Similar articles
-
Characterization of the breast cancer resistance protein (BCRP/ABCG2) in clear cell renal cell carcinoma.Int J Cancer. 2018 Dec 15;143(12):3181-3193. doi: 10.1002/ijc.31741. Epub 2018 Sep 25. Int J Cancer. 2018. PMID: 30070687
-
Stemness and anti-cancer drug resistance in ATP-binding cassette subfamily G member 2 highly expressed pancreatic cancer is induced in 3D culture conditions.Cancer Sci. 2018 Apr;109(4):1135-1146. doi: 10.1111/cas.13533. Epub 2018 Mar 25. Cancer Sci. 2018. PMID: 29444383 Free PMC article.
-
Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers.Comput Struct Biotechnol J. 2022 Jun 18;20:3106-3119. doi: 10.1016/j.csbj.2022.06.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782736 Free PMC article.
-
Multidrug efflux transporter ABCG2: expression and regulation.Cell Mol Life Sci. 2021 Nov;78(21-22):6887-6939. doi: 10.1007/s00018-021-03901-y. Epub 2021 Sep 29. Cell Mol Life Sci. 2021. PMID: 34586444 Free PMC article. Review.
-
Multidrug resistance in cancer chemotherapy and xenobiotic protection mediated by the half ATP-binding cassette transporter ABCG2.Curr Med Chem Anticancer Agents. 2004 Jan;4(1):31-42. doi: 10.2174/1568011043482205. Curr Med Chem Anticancer Agents. 2004. PMID: 14754410 Review.
Cited by
-
ABCG2 Gene Expression in Non-Small Cell Lung Cancer.Biomedicines. 2024 Oct 19;12(10):2394. doi: 10.3390/biomedicines12102394. Biomedicines. 2024. PMID: 39457707 Free PMC article.
-
ABCG2 Expression as a Potential Survival Predictor in Human Gliomas.Int J Mol Sci. 2024 Mar 8;25(6):3116. doi: 10.3390/ijms25063116. Int J Mol Sci. 2024. PMID: 38542090 Free PMC article.
-
ABCG2 protein expression in tumors of patients with non-resectable pancreatic cancer treated with gemcitabine and nab-paclitaxel.Front Oncol. 2025 Jun 6;15:1558184. doi: 10.3389/fonc.2025.1558184. eCollection 2025. Front Oncol. 2025. PMID: 40548120 Free PMC article.
-
Elacridar Inhibits BCRP Protein Activity in 2D and 3D Cell Culture Models of Ovarian Cancer and Re-Sensitizes Cells to Cytotoxic Drugs.Int J Mol Sci. 2025 Jun 17;26(12):5800. doi: 10.3390/ijms26125800. Int J Mol Sci. 2025. PMID: 40565261 Free PMC article.
-
PANTAX: a phase Ib clinical trial of the efflux pump inhibitor SCO-101 in combination with gemcitabine and nab-paclitaxel in non-resectable or metastatic pancreatic cancer.Invest New Drugs. 2025 Apr;43(2):337-347. doi: 10.1007/s10637-025-01526-7. Epub 2025 Apr 24. Invest New Drugs. 2025. PMID: 40272619 Free PMC article. Clinical Trial.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

