High-Resolution Small RNAs Landscape Provides Insights into Alkane Adaptation in the Marine Alkane-Degrader Alcanivorax dieselolei B-5
- PMID: 36555635
- PMCID: PMC9788540
- DOI: 10.3390/ijms232415995
High-Resolution Small RNAs Landscape Provides Insights into Alkane Adaptation in the Marine Alkane-Degrader Alcanivorax dieselolei B-5
Abstract
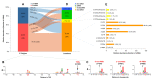
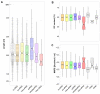
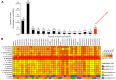
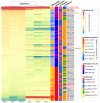
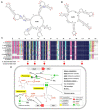
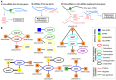
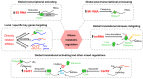
Alkanes are widespread in the ocean, and Alcanivorax is one of the most ubiquitous alkane-degrading bacteria in the marine ecosystem. Small RNAs (sRNAs) are usually at the heart of regulatory pathways, but sRNA-mediated alkane metabolic adaptability still remains largely unknown due to the difficulties of identification. Here, differential RNA sequencing (dRNA-seq) modified with a size selection (~50-nt to 500-nt) strategy was used to generate high-resolution sRNAs profiling in the model species Alcanivorax dieselolei B-5 under alkane (n-hexadecane) and non-alkane (acetate) conditions. As a result, we identified 549 sRNA candidates at single-nucleotide resolution of 5'-ends, 63.4% of which are with transcription start sites (TSSs), and 36.6% of which are with processing sites (PSSs) at the 5'-ends. These sRNAs originate from almost any location in the genome, regardless of intragenic (65.8%), antisense (20.6%) and intergenic (6.2%) regions, and RNase E may function in the maturation of sRNAs. Most sRNAs locally distribute across the 15 reference genomes of Alcanivorax, and only 7.5% of sRNAs are broadly conserved in this genus. Expression responses to the alkane of several core conserved sRNAs, including 6S RNA, M1 RNA and tmRNA, indicate that they may participate in alkane metabolisms and result in more actively global transcription, RNA processing and stresses mitigation. Two novel CsrA-related sRNAs are identified, which may be involved in the translational activation of alkane metabolism-related genes by sequestering the global repressor CsrA. The relationships of sRNAs with the characterized genes of alkane sensing (ompS), chemotaxis (mcp, cheR, cheW2), transporting (ompT1, ompT2, ompT3) and hydroxylation (alkB1, alkB2, almA) were created based on the genome-wide predicted sRNA-mRNA interactions. Overall, the sRNA landscape lays the ground for uncovering cryptic regulations in critical marine bacterium, among which both the core and species-specific sRNAs are implicated in the alkane adaptive metabolisms.
Keywords: Alcanivorax; PSS; TSS; alkane; dRNA-seq; metabolic regulation; sRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The long-chain alkane metabolism network of Alcanivorax dieselolei.Nat Commun. 2014 Dec 12;5:5755. doi: 10.1038/ncomms6755. Nat Commun. 2014. PMID: 25502912
-
Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5.Environ Microbiol. 2011 May;13(5):1168-78. doi: 10.1111/j.1462-2920.2010.02416.x. Epub 2011 Jan 24. Environ Microbiol. 2011. PMID: 21261799
-
Transcriptome landscape of Lactococcus lactis reveals many novel RNAs including a small regulatory RNA involved in carbon uptake and metabolism.RNA Biol. 2016;13(3):353-66. doi: 10.1080/15476286.2016.1146855. Epub 2016 Mar 7. RNA Biol. 2016. PMID: 26950529 Free PMC article.
-
Small RNA-mediated regulation in bacteria: A growing palette of diverse mechanisms.Gene. 2018 May 20;656:60-72. doi: 10.1016/j.gene.2018.02.068. Epub 2018 Mar 1. Gene. 2018. PMID: 29501814 Review.
-
Small RNAs in streptococci.RNA Biol. 2012 Apr;9(4):414-26. doi: 10.4161/rna.20104. Epub 2012 Apr 1. RNA Biol. 2012. PMID: 22546939 Review.
References
-
- Lea-Smith D.J., Biller S.J., Davey M.P., Cotton C.A., Perez Sepulveda B.M., Turchyn A.V., Scanlan D.J., Smith A.G., Chisholm S.W., Howe C.J. Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc. Natl. Acad. Sci. USA. 2015;112:13591–13596. doi: 10.1073/pnas.1507274112. - DOI - PMC - PubMed
-
- Scoma A., Yakimov M.M., Daffonchio D., Boon N. Self-healing capacity of deep-sea ecosystems affected by petroleum hydrocarbons: Understanding microbial oil degradation at hydrocarbon seeps is key to sustainable bioremediation protocols. EMBO Rep. 2017;18:868–872. doi: 10.15252/embr.201744090. - DOI - PMC - PubMed
-
- Gregson B.H., Metodieva G., Metodiev M.V., McKew B.A. Differential protein expression during growth on linear versus branched alkanes in the obligate marine hydrocarbon-degrading bacterium Alcanivorax borkumensis SK2T. Environ. Microbiol. 2019;21:2347–2359. doi: 10.1111/1462-2920.14620. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

