PLEKHM2 Loss of Function Impairs the Activity of iPSC-Derived Neurons via Regulation of Autophagic Flux
- PMID: 36555735
- PMCID: PMC9782635
- DOI: 10.3390/ijms232416092
PLEKHM2 Loss of Function Impairs the Activity of iPSC-Derived Neurons via Regulation of Autophagic Flux
Abstract
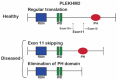
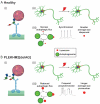
Pleckstrin Homology And RUN Domain Containing M2 (PLEKHM2) [delAG] mutation causes dilated cardiomyopathy with left ventricular non-compaction (DCM-LVNC), resulting in a premature death of PLEKHM2[delAG] individuals due to heart failure. PLEKHM2 is a factor involved in autophagy, a master regulator of cellular homeostasis, decomposing pathogens, proteins and other cellular components. Autophagy is mainly carried out by the lysosome, containing degradation enzymes, and by the autophagosome, which engulfs substances marked for decomposition. PLEKHM2 promotes lysosomal movement toward the cell periphery. Autophagic dysregulation is associated with neurodegenerative diseases' pathogenesis. Thus, modulation of autophagy holds considerable potential as a therapeutic target for such disorders. We hypothesized that PLEKHM2 is involved in neuronal development and function, and that mutated PLEKHM2 (PLEKHM2[delAG]) neurons will present impaired functions. Here, we studied PLEKHM2-related abnormalities in induced pluripotent stem cell (iPSC)-derived motor neurons (iMNs) as a neuronal model. PLEKHM2[delAG] iMN cultures had healthy control-like differentiation potential but exhibited reduced autophagic activity. Electrophysiological measurements revealed that PLEKHM2[delAG] iMN cultures displayed delayed functional maturation and more frequent and unsynchronized activity. This was associated with increased size and a more perinuclear lysosome cellular distribution. Thus, our results suggest that PLEKHM2 is involved in the functional development of neurons through the regulation of autophagic flux.
Keywords: DCM-LVNC; PLEKHM2; autophagosomes; autophagy; disease model; iPSCs; lysosomes; motor neurons; neurodegeneration; neurons.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Muhammad E., Levitas A., Singh S.R., Braiman A., Ofir R., Etzion S., Sheffield V.C., Etzion Y., Carrier L., Parvari R. PLEKHM2 Mutation Leads to Abnormal Localization of Lysosomes, Impaired Autophagy Flux and Associates with Recessive Dilated Cardiomyopathy and Left Ventricular Noncompaction. Hum. Mol. Genet. 2015;24:7227–7240. doi: 10.1093/hmg/ddv423. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

