A General Signal Pathway to Regulate Multiple Detoxification Genes Drives the Evolution of Helicoverpa armigera Adaptation to Xenobiotics
- PMID: 36555764
- PMCID: PMC9788003
- DOI: 10.3390/ijms232416126
A General Signal Pathway to Regulate Multiple Detoxification Genes Drives the Evolution of Helicoverpa armigera Adaptation to Xenobiotics
Abstract
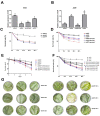
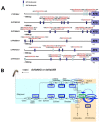
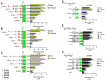
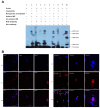
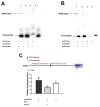
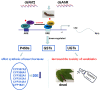
The study of insect adaptation to the defensive metabolites of host plants and various kinds of insecticides in order to acquire resistance is a hot topic in the pest-control field, but the mechanism is still unclear. In our study, we found that a general signal pathway exists in H. armigera which can regulate multiple P450s, GSTs and UGTs genes to help insects decrease their susceptibility to xenobiotics. Knockdown of HaNrf2 and HaAhR expression could significantly increase the toxicity of xenobiotics to H. armigera, and simultaneously decrease the gene expression of P450s, GSTs and UGTs which are related to the xenobiotic metabolism and synthesis of insect hormone pathways. Then, we used EMSA and dual luciferase assay to verify that a crosstalk exists between AhR and Nrf2 to regulate multiple P450s, GSTs and UGTs genes to mediate H. armigera susceptibility to plant allelochemicals and insecticides. The detoxification genes' expression network which can be regulated by Nrf2 and AhR is still unknown, and there were also no reports about the crosstalk between AhR and Nrf2 that exist in insects and can regulate multiple detoxification genes' expression. Our results provide a new general signaling pathway to reveal the adaptive mechanism of insects to xenobiotics and provides further insight into designing effective pest-management strategies to avoid the overuse of insecticides.
Keywords: detoxification genes; regulation; transcription factor; xenobiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Effects of different insecticides on transcripts of key genes in CncC pathway and detoxification genes in Helicoverpa armigera.Pestic Biochem Physiol. 2023 Sep;195:105541. doi: 10.1016/j.pestbp.2023.105541. Epub 2023 Jul 19. Pestic Biochem Physiol. 2023. PMID: 37666612
-
Overexpression of the F116V allele of CYP9A186 in transgenic Helicoverpa armigera confers high-level resistance to emamectin benzoate.Insect Biochem Mol Biol. 2023 Dec;163:104042. doi: 10.1016/j.ibmb.2023.104042. Epub 2023 Nov 27. Insect Biochem Mol Biol. 2023. PMID: 38030045
-
Involvement of CYP2 and mitochondrial clan P450s of Helicoverpa armigera in xenobiotic metabolism.Insect Biochem Mol Biol. 2022 Jan;140:103696. doi: 10.1016/j.ibmb.2021.103696. Epub 2021 Nov 17. Insect Biochem Mol Biol. 2022. PMID: 34800643
-
Xenobiotic responses in insects.Arch Insect Biochem Physiol. 2022 Mar;109(3):e21869. doi: 10.1002/arch.21869. Epub 2022 Jan 28. Arch Insect Biochem Physiol. 2022. PMID: 35088911 Review.
-
The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics.Curr Opin Insect Sci. 2021 Feb;43:103-107. doi: 10.1016/j.cois.2020.11.004. Epub 2020 Dec 30. Curr Opin Insect Sci. 2021. PMID: 33387688 Review.
Cited by
-
A detoxification pathway initiated by a nuclear receptor TcHR96h in Tetranychus cinnabarinus (Boisduval).PLoS Genet. 2023 Sep 14;19(9):e1010911. doi: 10.1371/journal.pgen.1010911. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37708138 Free PMC article.
-
Indole-3-Lactic Acid Inhibits Doxorubicin-Induced Ferroptosis Through Activating Aryl Hydrocarbon Receptor/Nrf2 Signalling Pathway.J Cell Mol Med. 2025 Jan;29(2):e70358. doi: 10.1111/jcmm.70358. J Cell Mol Med. 2025. PMID: 39854052 Free PMC article.
-
Self-assembled co-delivery nanoplatform for increasing the broad-spectrum susceptibility of fall armyworm toward insecticides.J Adv Res. 2025 Jan;67:93-104. doi: 10.1016/j.jare.2024.01.031. Epub 2024 Jan 28. J Adv Res. 2025. PMID: 38286302 Free PMC article.
References
-
- Heckel D.G. Annu. Plant Rev. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2014. Insect detoxification and sequestration strategies; pp. 77–114.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

