Effect of Superovulation Treatment on Oocyte's DNA Methylation
- PMID: 36555801
- PMCID: PMC9785075
- DOI: 10.3390/ijms232416158
Effect of Superovulation Treatment on Oocyte's DNA Methylation
Abstract
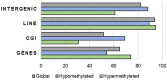
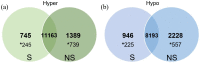
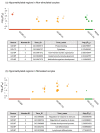
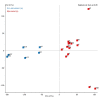
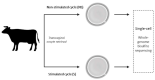
Controlled ovarian stimulation is a necessary step in some assisted reproductive procedures allowing a higher collection of female gametes. However, consequences of this stimulation for the gamete or the offspring have been shown in several mammals. Most studies used comparisons between oocytes from different donors, which may contribute to different responses. In this work, we use the bovine model in which each animal serves as its own control. DNA methylation profiles were obtained by single-cell whole-genome bisulfite sequencing of oocytes from pre-ovulatory unstimulated follicles compared to oocytes from stimulated follicles. Results show that the global percentage of methylation was similar between groups, but the percentage of methylation was lower for non-stimulated oocytes in the imprinted genes APEG3, MEG3, and MEG9 and higher in TSSC4 when compared to stimulated oocytes. Differences were also found in CGI of imprinted genes: higher methylation was found among non-stimulated oocytes in MEST (PEG1), IGF2R, GNAS (SCG6), KvDMR1 ICR UMD, and IGF2. In another region around IGF2, the methylation percentage was lower for non-stimulated oocytes when compared to stimulated oocytes. Data drawn from this study might help to understand the molecular reasons for the appearance of certain syndromes in assisted reproductive technologies-derived offspring.
Keywords: ART; DNA methylation; epigenetics; oocyte; ovarian stimulation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

