Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma
- PMID: 36556220
- PMCID: PMC9785654
- DOI: 10.3390/jpm12121999
Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma
Abstract
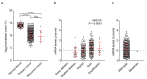
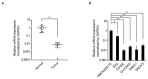
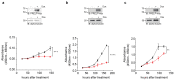
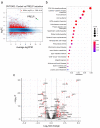
Ovarian clear cell carcinoma (OCCC) has a poor prognosis, and its therapeutic strategy has not been established. PRELP is a leucine-rich repeat protein in the extracellular matrix of connective tissues. Although PRELP anchors the basement membrane to the connective tissue and is absent in most epithelial cancers, much remains unknown regarding its function as a regulator of ligand-mediated signaling pathways. Here, we obtained sets of differentially expressed genes by PRELP expression using OCCC cell lines. We found that more than 1000 genes were significantly altered by PRELP expression, particularly affecting the expression of a group of genes involved in the PI3K-AKT signaling pathway. Furthermore, we revealed the loss of active histone marks on the loci of the PRELP gene in patients with OCCC and how its forced expression inhibited cell proliferation. These findings suggest that PRELP is not only a molecule anchored in connective tissues but is also a signaling molecule acting in a tumor-suppressive manner. It can serve as the basis for early detection and novel therapeutic approaches for OCCC toward precision medicine.
Keywords: PRELP; epigenetics; gene expression; ovarian clear cell carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The leucine-rich repeat protein PRELP binds perlecan and collagens and may function as a basement membrane anchor.J Biol Chem. 2002 Apr 26;277(17):15061-8. doi: 10.1074/jbc.M108285200. Epub 2002 Feb 14. J Biol Chem. 2002. PMID: 11847210
-
Proteomic identification and validation of novel interactions of the putative tumor suppressor PRELP with membrane proteins including IGFI-R and p75NTR.J Biol Chem. 2021 Jan-Jun;296:100278. doi: 10.1016/j.jbc.2021.100278. Epub 2021 Jan 9. J Biol Chem. 2021. PMID: 33428936 Free PMC article.
-
Repression of the PRELP gene is relieved by histone deacetylase inhibitors through acetylation of histone H2B lysine 5 in bladder cancer.Clin Epigenetics. 2022 Nov 12;14(1):147. doi: 10.1186/s13148-022-01370-z. Clin Epigenetics. 2022. PMID: 36371227 Free PMC article.
-
PRELP, collagen, and a theory of Hutchinson-Gilford progeria.Ageing Res Rev. 2003 Jan;2(1):95-105. doi: 10.1016/s1568-1637(02)00044-2. Ageing Res Rev. 2003. PMID: 12437997 Review.
-
Updates of Pathogenesis, Diagnostic and Therapeutic Perspectives for Ovarian Clear Cell Carcinoma.J Cancer. 2021 Feb 22;12(8):2295-2316. doi: 10.7150/jca.53395. eCollection 2021. J Cancer. 2021. PMID: 33758607 Free PMC article. Review.
Cited by
-
Investigating the role of disulfidptosis related genes in radiotherapy resistance of lung adenocarcinoma.Front Med (Lausanne). 2024 Oct 23;11:1473080. doi: 10.3389/fmed.2024.1473080. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39507711 Free PMC article.
-
PRELP regulated by GAS5/miR-3127-5p suppresses cisplatin resistance in oral squamous cell carcinoma.Cytotechnology. 2025 Jun;77(3):92. doi: 10.1007/s10616-025-00749-z. Epub 2025 Apr 28. Cytotechnology. 2025. PMID: 40309012
-
Correlation of the tumor escape phenotype with loss of PRELP expression in melanoma.J Transl Med. 2023 Sep 20;21(1):643. doi: 10.1186/s12967-023-04476-x. J Transl Med. 2023. PMID: 37730606 Free PMC article.
-
Impact of dcEF on microRNA profiles in glioblastoma and exosomes using a novel microfluidic bioreactor.Biomicrofluidics. 2024 Dec 27;18(6):064106. doi: 10.1063/5.0228901. eCollection 2024 Dec. Biomicrofluidics. 2024. PMID: 39742343 Free PMC article.
-
Comprehensive investigation of proteoglycan gene expression in breast cancer: Discovery of a unique proteoglycan gene signature linked to the malignant phenotype.Proteoglycan Res. 2025 Jan-Mar;3(1):e70014. doi: 10.1002/pgr2.70014. Epub 2025 Jan 8. Proteoglycan Res. 2025. PMID: 40066261 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

