Novel Virus Identification through Metagenomics: A Systematic Review
- PMID: 36556413
- PMCID: PMC9784588
- DOI: 10.3390/life12122048
Novel Virus Identification through Metagenomics: A Systematic Review
Abstract
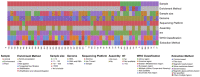
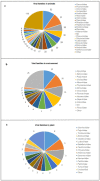
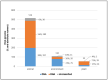
Metagenomic Next Generation Sequencing (mNGS) allows the evaluation of complex microbial communities, avoiding isolation and cultivation of each microbial species, and does not require prior knowledge of the microbial sequences present in the sample. Applications of mNGS include virome characterization, new virus discovery and full-length viral genome reconstruction, either from virus preparations enriched in culture or directly from clinical and environmental specimens. Here, we systematically reviewed studies that describe novel virus identification through mNGS from samples of different origin (plant, animal and environment). Without imposing time limits to the search, 379 publications were identified that met the search parameters. Sample types, geographical origin, enrichment and nucleic acid extraction methods, sequencing platforms, bioinformatic analytical steps and identified viral families were described. The review highlights mNGS as a feasible method for novel virus discovery from samples of different origins, describes which kind of heterogeneous experimental and analytical protocols are currently used and provides useful information such as the different commercial kits used for the purification of nucleic acids and bioinformatics analytical pipelines.
Keywords: NGS; animal; environment; novel virus; plant; viral metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

