Characterization of Mixed-Species Biofilms Formed by Four Gut Microbiota
- PMID: 36557585
- PMCID: PMC9781930
- DOI: 10.3390/microorganisms10122332
Characterization of Mixed-Species Biofilms Formed by Four Gut Microbiota
Abstract
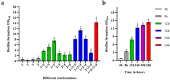
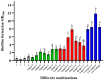
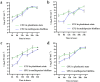
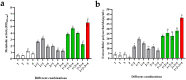
In natural settings, approximately 40-80% of bacteria exist as biofilms, most of which are mixed-species biofilms. Previous studies have typically focused on single- or dual-species biofilms. To expand the field of study on gut biofilms, we found a group of gut microbiota that can form biofilms well in vitro: Bifidobacterium longum subsp. infantis, Enterococcus faecalis, Bacteroides ovatus, and Lactobacillus gasseri. The increase in biomass and bio-volume of the mixed-species biofilm was confirmed via crystal violet staining, field emission scanning electron microscopy, and confocal laser scanning microscopy, revealing a strong synergistic relationship in these communities, with B. longum being the key biofilm-contributing species. This interaction may be related to changes in the cell number, biofilm-related genes, and metabolic activities. After quantifying the cell number using quantitative polymerase chain reaction, B. longum and L. gasseri were found to be the dominant flora in the mixed-species biofilm. In addition, this study analyzed biological properties of mixed-species biofilms, such as antibiotic resistance, cell metabolic activity, and concentration of water-insoluble polysaccharides. Compared with single-species biofilms, mixed-species biofilms had higher metabolic activity, more extracellular matrix, and greater antibiotic resistance. From these results, we can see that the formation of biofilms is a self-protection mechanism of gut microbiota, and the formation of mixed-species biofilms can greatly improve the survival rate of different strains. Finally, this study is a preliminary exploration of the biological characteristics of gut biofilms, and the molecular mechanisms underlying the formation of biofilms warrant further research.
Keywords: Bacteroides; Bifidobacterium; Enterococcus; Lactobacillus; gut microbiota; mixed-species biofilms; synergistic interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Synergistic interactions prevail in multispecies biofilms formed by the human gut microbiota on mucin.FEMS Microbiol Ecol. 2021 Jul 13;97(8):fiab096. doi: 10.1093/femsec/fiab096. FEMS Microbiol Ecol. 2021. PMID: 34190973
-
Transcriptional Changes in Bifidobacterium bifidum Involved in Synergistic Multispecies Biofilms.Microb Ecol. 2022 Oct;84(3):922-934. doi: 10.1007/s00248-021-01904-7. Epub 2021 Oct 21. Microb Ecol. 2022. PMID: 34676439
-
Multiomics reveals the mechanism of B. longum in promoting the formation of mixed-species biofilms.Food Funct. 2023 Sep 19;14(18):8276-8290. doi: 10.1039/d3fo01751f. Food Funct. 2023. PMID: 37602484
-
Inhibitory effect of probiotic lactobacilli supernatants on single and mixed non-albicans Candida species biofilm.Arch Oral Biol. 2018 Jan;85:40-45. doi: 10.1016/j.archoralbio.2017.10.002. Epub 2017 Oct 6. Arch Oral Biol. 2018. PMID: 29031236
-
Gut biofilms: Bacteroides as model symbionts to study biofilm formation by intestinal anaerobes.FEMS Microbiol Rev. 2022 Mar 3;46(2):fuab054. doi: 10.1093/femsre/fuab054. FEMS Microbiol Rev. 2022. PMID: 34849798 Review.
Cited by
-
Medical Device-Associated Infections Caused by Biofilm-Forming Microbial Pathogens and Controlling Strategies.Antibiotics (Basel). 2024 Jul 4;13(7):623. doi: 10.3390/antibiotics13070623. Antibiotics (Basel). 2024. PMID: 39061305 Free PMC article. Review.
-
Cyanobacteria and Chloroflexota cooperate to structure light-responsive biofilms.Proc Natl Acad Sci U S A. 2025 Feb 4;122(5):e2423574122. doi: 10.1073/pnas.2423574122. Epub 2025 Jan 29. Proc Natl Acad Sci U S A. 2025. PMID: 39879238 Free PMC article.
-
In Vitro Inhibitory Effect of Silver Diamine Fluoride Combined with Potassium Iodide against Mixed-Species Biofilm Formation on Human Root Dentin.Antibiotics (Basel). 2024 Aug 7;13(8):743. doi: 10.3390/antibiotics13080743. Antibiotics (Basel). 2024. PMID: 39200043 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

