The Spatial Organization of Bacterial Transcriptional Regulatory Networks
- PMID: 36557619
- PMCID: PMC9787925
- DOI: 10.3390/microorganisms10122366
The Spatial Organization of Bacterial Transcriptional Regulatory Networks
Abstract
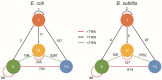
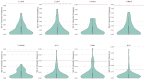
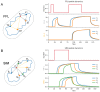
The transcriptional regulatory network (TRN) is the central pivot of a prokaryotic organism to receive, process and respond to internal and external environmental information. However, little is known about its spatial organization so far. In recent years, chromatin interaction data of bacteria such as Escherichia coli and Bacillus subtilis have been published, making it possible to study the spatial organization of bacterial transcriptional regulatory networks. By combining TRNs and chromatin interaction data of E. coli and B. subtilis, we explored the spatial organization characteristics of bacterial TRNs in many aspects such as regulation directions (positive and negative), central nodes (hubs, bottlenecks), hierarchical levels (top, middle, bottom) and network motifs (feed-forward loops and single input modules) of the TRNs and found that the bacterial TRNs have a variety of stable spatial organization features under different physiological conditions that may be closely related with biological functions. Our findings provided new insights into the connection between transcriptional regulation and the spatial organization of chromosome in bacteria and might serve as a factual foundation for trying spatial-distance-based gene circuit design in synthetic biology.
Keywords: 3D genome; bacterial chromatin; network hierarchy; network motif; spatial effect.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Balleza E., Lopez-Bojorquez L.N., Martinez-Antonio A., Resendis-Antonio O., Lozada-Chavez I., Balderas-Martinez Y.I., Encarnacion S., Collado-Vides J. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol. Rev. 2009;33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

