Generation and Utilization of a Monoclonal Antibody against Hepatitis B Virus Core Protein for a Comprehensive Interactome Analysis
- PMID: 36557634
- PMCID: PMC9783060
- DOI: 10.3390/microorganisms10122381
Generation and Utilization of a Monoclonal Antibody against Hepatitis B Virus Core Protein for a Comprehensive Interactome Analysis
Abstract
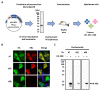
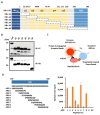
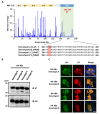
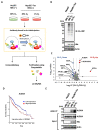
Hepatitis B virus (HBV) core antigen (HBc) is a structural protein that forms the viral nucleocapsid and is involved in various steps of the viral replication cycle, but its role in the pathogenesis of HBV infection is still elusive. In this study, we generated a mouse monoclonal antibody (mAb) against HBc and used it in antibody-based in situ biotinylation analysis in order to identify host proteins that interact with HBc. HBc antigen was produced with a wheat germ cell-free protein synthesis system and used to immunize mice. Among the established hybridoma clones, a single clone (mAb #7) was selected and further characterized for its ability in the antibody-based in situ biotinylation analysis to collect host proteins that are in the vicinity of HBc. Using mass spectrometry, we identified 215 HBc-interacting host proteins, three of which bind HBc most significantly under hypoxic conditions. Our results indicate that mAb #7 can be used to systematically identify host proteins that interact with HBc under pathophysiological conditions, and thus may be useful to explore the molecular pathways involved in HBV-induced cytopathogenesis.
Keywords: Hepatitis B virus; antibody-based in situ biotinylation; hypoxia; interactome analysis; monoclonal antibody.
Conflict of interest statement
Y.Y. is a current employee of Kanto Chemical Co., Inc. A.R. received a collaborative research grant from Kanto Chemical Co., Inc.
Figures

Similar articles
-
Intracellular single-chain antibody against hepatitis B virus core protein inhibits the replication of hepatitis B virus in cultured cells.Hepatology. 1999 Jul;30(1):300-7. doi: 10.1002/hep.510300105. Hepatology. 1999. PMID: 10385671
-
Hepatitis B virus core antigen (HBc Ag) accumulation in an HBV nonproducer clone of HepG2-transfected cells is associated with cytopathic effect.Virology. 1990 Nov;179(1):113-20. doi: 10.1016/0042-6822(90)90280-5. Virology. 1990. PMID: 2171201
-
Regulation of Hepatitis B Virus Replication by Cyclin Docking Motifs in Core Protein.J Virol. 2021 May 24;95(12):e00230-21. doi: 10.1128/JVI.00230-21. Print 2021 May 24. J Virol. 2021. PMID: 33789995 Free PMC article.
-
The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: Implications for the development of HBc-targeting antivirals.Antiviral Res. 2018 Jan;149:211-220. doi: 10.1016/j.antiviral.2017.11.015. Epub 2017 Nov 26. Antiviral Res. 2018. PMID: 29183719 Free PMC article. Review.
-
A Pleiotropic Role of the Hepatitis B Virus Core Protein in Hepatocarcinogenesis.Int J Mol Sci. 2021 Dec 20;22(24):13651. doi: 10.3390/ijms222413651. Int J Mol Sci. 2021. PMID: 34948447 Free PMC article. Review.
Cited by
-
The characterization and structural basis of a human broadly binding antibody to HBV core protein.J Virol. 2025 Jan 31;99(1):e0169424. doi: 10.1128/jvi.01694-24. Epub 2024 Nov 27. J Virol. 2025. PMID: 39601613 Free PMC article.
References
-
- D’Arienzo V., Ferguson J., Giraud G., Chapus F., Harris J.M., Wing P.A.C., Claydon A., Begum S., Zhuang X., Balfe P., et al. The CCCTC-binding factor CTCF represses Hepatitis B virus enhancer I and regulates viral transcription. Cell. Microbiol. 2021;23:e13274. doi: 10.1111/cmi.13274. - DOI - PMC - PubMed
-
- Virzi A., Motos V.G., Tripon S., Baumert T.F., Virzi A., Motos V.G., Tripon S., Baumert T.F., Profi- J.L., Virz A., et al. Profibrotic Signaling and HCC Risk during Chronic Viral Hepatitis: Biomarker Development To cite this version: HAL Id: Hal-03603544 Profibrotic Signaling and HCC Risk during Chronic Viral Hepatitis: Biomarker Development. J. Clin. Med. 2021;10:977. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

