Safe Sialidase Production by the Saprophyte Oerskovia paurometabola: Gene Sequence and Enzyme Purification
- PMID: 36558051
- PMCID: PMC9782813
- DOI: 10.3390/molecules27248922
Safe Sialidase Production by the Saprophyte Oerskovia paurometabola: Gene Sequence and Enzyme Purification
Abstract
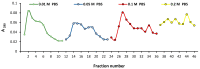
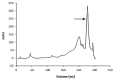
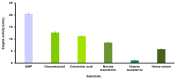
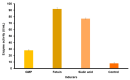
Sialidase preparations are applied in structural and functional studies on sialoglycans, in the production of sialylated therapeutic proteins and synthetic substrates for use in biochemical research, etc. They are obtained mainly from pathogenic microorganisms; therefore, the demand for apathogenic producers of sialidase is of exceptional importance for the safe production of this enzyme. Here, we report for the first time the presence of a sialidase gene and enzyme in the saprophytic actinomycete Oerskovia paurometabola strain O129. An electrophoretically pure, glycosylated enzyme with a molecular weight of 70 kDa was obtained after a two-step chromatographic procedure using DEAE cellulose and Q-sepharose. The biochemical characterization showed that the enzyme is extracellular, inductive, and able to cleave α(2→3,6,8) linked sialic acids with preference for α(2→3) bonds. The enzyme production was strongly induced by glycomacropeptide (GMP) from milk whey, as well as by sialic acid. Investigation of the deduced amino acid sequence revealed that the protein molecule has the typical six-bladed β-propeller structure and contains all features of bacterial sialidases, i.e., an YRIP motif, five Asp-boxes, and the conserved amino acids in the active site. The presence of an unusual signal peptide of 40 amino acids was predicted. The sialidase-producing O. paurometabola O129 showed high and constant enzyme production. Together with its saprophytic nature, this makes it a reliable producer with high potential for industrial application.
Keywords: Oerskovia paurometabola; enzyme purification; gene sequencing; sialidase.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Novel sialidase from non-pathogenic bacterium Oerskovia paurometabola strain O129.Z Naturforsch C J Biosci. 2022 Nov 10;78(1-2):49-55. doi: 10.1515/znc-2022-0051. Print 2023 Jan 27. Z Naturforsch C J Biosci. 2022. PMID: 36351238
-
Cloning and characterization of a sialidase from the filamentous fungus, Aspergillus fumigatus.Glycoconj J. 2010 Jul;27(5):533-48. doi: 10.1007/s10719-010-9299-9. Epub 2010 Jul 23. Glycoconj J. 2010. PMID: 20652740
-
Structural studies on the Pseudomonas aeruginosa sialidase-like enzyme PA2794 suggest substrate and mechanistic variations.J Mol Biol. 2009 Feb 27;386(3):828-40. doi: 10.1016/j.jmb.2008.12.084. Epub 2009 Jan 10. J Mol Biol. 2009. PMID: 19166860
-
Comparative enzymology, biochemistry and pathophysiology of human exo-alpha-sialidases (neuraminidases).Comp Biochem Physiol B Biochem Mol Biol. 2001 May;129(1):29-64. doi: 10.1016/s1096-4959(01)00372-4. Comp Biochem Physiol B Biochem Mol Biol. 2001. PMID: 11337249 Review.
-
Trans-sialidase: a unique enzyme activity discovered in the protozoan Trypanosoma cruzi.FASEB J. 1993 Oct;7(13):1257-64. doi: 10.1096/fasebj.7.13.8405811. FASEB J. 1993. PMID: 8405811 Review.
Cited by
-
Structural and functional characterization of cold-active sialidase isolated from Antarctic fungus Penicillium griseofulvum P29.Biochem Biophys Rep. 2023 Dec 20;37:101610. doi: 10.1016/j.bbrep.2023.101610. eCollection 2024 Mar. Biochem Biophys Rep. 2023. PMID: 38155944 Free PMC article.
References
-
- Eneva R., Engibarov S., Abrashev R., Krumova E., Angelova M. Sialic acids, sialoconjugates and enzymes of their metabolism in fungi. Biotechnol. Biotechnol. Equip. 2021;35:346–357. doi: 10.1080/13102818.2021.1879678. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

