Short Chain Fatty Acid Metabolism in Relation to Gut Microbiota and Genetic Variability
- PMID: 36558520
- PMCID: PMC9788597
- DOI: 10.3390/nu14245361
Short Chain Fatty Acid Metabolism in Relation to Gut Microbiota and Genetic Variability
Abstract
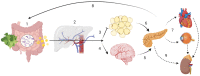
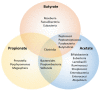
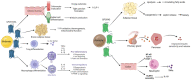
It is widely accepted that the gut microbiota plays a significant role in modulating inflammatory and immune responses of their host. In recent years, the host-microbiota interface has gained relevance in understanding the development of many non-communicable chronic conditions, including cardiovascular disease, cancer, autoimmunity and neurodegeneration. Importantly, dietary fibre (DF) and associated compounds digested by the microbiota and their resulting metabolites, especially short-chain fatty acids (SCFA), were significantly associated with health beneficial effects, such as via proposed anti-inflammatory mechanisms. However, SCFA metabolic pathways are not fully understood. Major steps include production of SCFA by microbiota, uptake in the colonic epithelium, first-pass effects at the liver, followed by biodistribution and metabolism at the host's cellular level. As dietary patterns do not affect all individuals equally, the host genetic makeup may play a role in the metabolic fate of these metabolites, in addition to other factors that might influence the microbiota, such as age, birth through caesarean, medication intake, alcohol and tobacco consumption, pathogen exposure and physical activity. In this article, we review the metabolic pathways of DF, from intake to the intracellular metabolism of fibre-derived products, and identify possible sources of inter-individual variability related to genetic variation. Such variability may be indicative of the phenotypic flexibility in response to diet, and may be predictive of long-term adaptations to dietary factors, including maladaptation and tissue damage, which may develop into disease in individuals with specific predispositions, thus allowing for a better prediction of potential health effects following personalized intervention with DF.
Keywords: SNPs; dietary fibre; holobiont; microbiome; nutrigenetics; nutrigenomics; short chain fatty acids; sustainable development; synergies; translational research.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Dietary glycation compounds - implications for human health.Crit Rev Toxicol. 2024 Sep;54(8):485-617. doi: 10.1080/10408444.2024.2362985. Epub 2024 Aug 16. Crit Rev Toxicol. 2024. PMID: 39150724
-
Gut microbiota determines the fate of dietary fiber-targeted interventions in host health.Gut Microbes. 2024 Jan-Dec;16(1):2416915. doi: 10.1080/19490976.2024.2416915. Epub 2024 Oct 17. Gut Microbes. 2024. PMID: 39418223 Free PMC article. Review.
-
The relationship between gut microbiota, short-chain fatty acids and type 2 diabetes mellitus: the possible role of dietary fibre.Acta Diabetol. 2021 Sep;58(9):1131-1138. doi: 10.1007/s00592-021-01727-5. Epub 2021 May 10. Acta Diabetol. 2021. PMID: 33970303 Free PMC article. Review.
-
Gut microbiota and energy balance: role in obesity.Proc Nutr Soc. 2015 Aug;74(3):227-34. doi: 10.1017/S0029665114001700. Epub 2014 Dec 18. Proc Nutr Soc. 2015. PMID: 25518735
-
Short-chain fatty acids as a link between diet and cardiometabolic risk: a narrative review.Lipids Health Dis. 2023 Mar 14;22(1):40. doi: 10.1186/s12944-023-01803-5. Lipids Health Dis. 2023. PMID: 36915164 Free PMC article. Review.
Cited by
-
Combined Effects of Metals, PCBs, Dioxins, and Furans on Cardiovascular Dysfunction.J Xenobiot. 2025 Jun 19;15(3):94. doi: 10.3390/jox15030094. J Xenobiot. 2025. PMID: 40558877 Free PMC article.
-
Association of a newly proposed dietary index for gut microbiota with phenotypic age acceleration: a cross-sectional study of NHANES 1999-2018.J Health Popul Nutr. 2025 Jul 12;44(1):251. doi: 10.1186/s41043-025-01007-w. J Health Popul Nutr. 2025. PMID: 40652280 Free PMC article.
-
Machine learning integrates region-specific microbial signatures to distinguish geographically adjacent populations within a province.Front Microbiol. 2025 Jul 11;16:1586195. doi: 10.3389/fmicb.2025.1586195. eCollection 2025. Front Microbiol. 2025. PMID: 40718818 Free PMC article.
-
The Efficacy of Yeast Supplementation on Monogastric Animal Performance-A Short Review.Life (Basel). 2023 Oct 11;13(10):2037. doi: 10.3390/life13102037. Life (Basel). 2023. PMID: 37895419 Free PMC article. Review.
-
The Association of Short-Chain Fatty Acids with the Occurrence of Gastrointestinal Symptoms in Infants.Int J Mol Sci. 2024 Nov 21;25(23):12487. doi: 10.3390/ijms252312487. Int J Mol Sci. 2024. PMID: 39684199 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

