[PRION+] States Are Associated with Specific Histone H3 Post-Translational Modification Changes
- PMID: 36558770
- PMCID: PMC9786042
- DOI: 10.3390/pathogens11121436
[PRION+] States Are Associated with Specific Histone H3 Post-Translational Modification Changes
Abstract
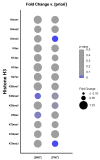
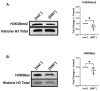
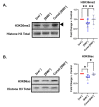
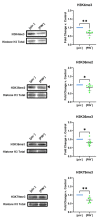
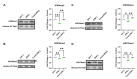
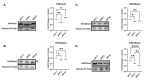
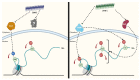
Prions are proteins able to take on alternative conformations and propagate them in a self-templating process. In Saccharomyces cerevisiae, prions enable heritable responses to environmental conditions through bet-hedging mechanisms. Hence, [PRION+] states may serve as an atypical form of epigenetic control, producing heritable phenotypic change via protein folding. However, the connections between prion states and the epigenome remain unknown. Do [PRION+] states link to canonical epigenetic channels, such as histone post-translational modifications? Here, we map out the histone H3 modification landscape in the context of the [SWI+] and [PIN+] prion states. [SWI+] is propagated by Swi1, a subunit of the SWI/SNF chromatin remodeling complex, while [PIN+] is propagated by Rnq1, a protein of unknown function. We find [SWI+] yeast display decreases in the levels of H3K36me2 and H3K56ac compared to [swi-] yeast. In contrast, decreases in H3K4me3, H3K36me2, H3K36me3 and H3K79me3 are connected to the [PIN+] state. Curing of the prion state by treatment with guanidine hydrochloride restored histone PTM to [prion-] state levels. We find histone PTMs in the [PRION+] state do not match those in loss-of-function models. Our findings shed light into the link between prion states and histone modifications, revealing novel insight into prion function in yeast.
Keywords: Saccharomyces cerevisiae; epigenetics; histone post-translational modifications; prion; protein aggregates; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

