Isolation and Characterization of Chi-like Salmonella Bacteriophages Infecting Two Salmonella enterica Serovars, Typhimurium and Enteritidis
- PMID: 36558814
- PMCID: PMC9783114
- DOI: 10.3390/pathogens11121480
Isolation and Characterization of Chi-like Salmonella Bacteriophages Infecting Two Salmonella enterica Serovars, Typhimurium and Enteritidis
Abstract
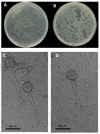
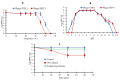
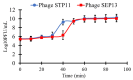
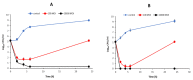
Salmonella enterica Serovar Typhimurium and Salmonella enterica Serovar Enteritidis are well-known pathogens that cause foodborne diseases in humans. The emergence of antibiotic-resistant Salmonella serovars has caused serious public health problems worldwide. In this study, two lysogenic phages, STP11 and SEP13, were isolated from a wastewater treatment plant in Jeddah, KSA. Transmission electron microscopic images revealed that both phages are new members of the genus “Chivirus” within the family Siphoviridae. Both STP11 and SEP13 had a lysis time of 90 min with burst sizes of 176 and 170 PFU/cell, respectively. The two phages were thermostable (0 °C ≤ temperature < 70 °C) and pH tolerant at 3 ≤ pH < 11. STP11 showed lytic activity for approximately 42.8% (n = 6), while SEP13 showed against 35.7% (n = 5) of the tested bacterial strains. STP11 and STP13 have linear dsDNA genomes consisting of 58,890 bp and 58,893 bp nucleotide sequences with G + C contents of 57% and 56.5%, respectively. Bioinformatics analysis revealed that the genomes of phages STP11 and SEP13 contained 70 and 71 ORFs, respectively. No gene encoding tRNA was detected in their genome. Of the 70 putative ORFs of phage STP11, 27 (38.6%) were assigned to functional genes and 43 (61.4%) were annotated as hypothetical proteins. Similarly, 29 (40.8%) of the 71 putative ORFs of phage SEP13 were annotated as functional genes, whereas the remaining 42 (59.2%) were assigned as nonfunctional proteins. Phylogenetic analysis of the whole genome sequence demonstrated that the isolated phages are closely related to Chi-like Salmonella viruses.
Keywords: Chi-like phages; S. Enteritidis; S. Typhimurium; bacteriophage; molecular characterization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Genomic and functional characterization of five novel Salmonella-targeting bacteriophages.Virol J. 2021 Sep 8;18(1):183. doi: 10.1186/s12985-021-01655-4. Virol J. 2021. PMID: 34496915 Free PMC article.
-
Isolation and characterization of polyvalent bacteriophages infecting multi drug resistant Salmonella serovars isolated from broilers in Egypt.Int J Food Microbiol. 2018 Feb 2;266:8-13. doi: 10.1016/j.ijfoodmicro.2017.11.009. Epub 2017 Nov 14. Int J Food Microbiol. 2018. PMID: 29156244
-
A Highly Effective Bacteriophage-1252 to Control Multiple Serovars of Salmonella enterica.Foods. 2023 Feb 13;12(4):797. doi: 10.3390/foods12040797. Foods. 2023. PMID: 36832872 Free PMC article.
-
Lysis Profiles of Salmonella Phages on Salmonella Isolates from Various Sources and Efficiency of a Phage Cocktail against S. Enteritidis and S. Typhimurium.Microorganisms. 2019 Apr 5;7(4):100. doi: 10.3390/microorganisms7040100. Microorganisms. 2019. PMID: 30959743 Free PMC article.
-
Limitation of the Lytic Effect of Bacteriophages on Salmonella and Other Enteric Bacterial Pathogens and Approaches to Overcome.Int J Microbiol. 2025 May 15;2025:5936070. doi: 10.1155/ijm/5936070. eCollection 2025. Int J Microbiol. 2025. PMID: 40405891 Free PMC article. Review.
Cited by
-
Evaluation of phage-antibiotic combinations in the treatment of extended-spectrum β-lactamase-producing Salmonella enteritidis strain PT1.Heliyon. 2023 Jan 19;9(1):e13077. doi: 10.1016/j.heliyon.2023.e13077. eCollection 2023 Jan. Heliyon. 2023. PMID: 36747932 Free PMC article.
-
Application of a novel lytic Jerseyvirus phage LPSent1 for the biological control of the multidrug-resistant Salmonella Enteritidis in foods.Front Microbiol. 2023 Apr 5;14:1135806. doi: 10.3389/fmicb.2023.1135806. eCollection 2023. Front Microbiol. 2023. PMID: 37089535 Free PMC article.
-
The Biotechnological Application of Bacteriophages: What to Do and Where to Go in the Middle of the Post-Antibiotic Era.Microorganisms. 2023 Sep 13;11(9):2311. doi: 10.3390/microorganisms11092311. Microorganisms. 2023. PMID: 37764155 Free PMC article. Review.
-
Isolation and Characterization of a Novel Lytic Phage, vB_PseuP-SA22, and Its Efficacy against Carbapenem-Resistant Pseudomonas aeruginosa.Antibiotics (Basel). 2023 Mar 2;12(3):497. doi: 10.3390/antibiotics12030497. Antibiotics (Basel). 2023. Retraction in: Antibiotics (Basel). 2024 Aug 28;13(9):814. doi: 10.3390/antibiotics13090814. PMID: 36978364 Free PMC article. Retracted.
-
Isolation and Characterization of a Bacteriophage with Potential for the Control of Multidrug-Resistant Salmonella Strains Encoding Virulence Factors Associated with the Promotion of Precancerous Lesions.Viruses. 2024 Oct 31;16(11):1711. doi: 10.3390/v16111711. Viruses. 2024. PMID: 39599826 Free PMC article.
References
-
- Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., International Collaboration on Enteric Disease “Burden of Illness” Studies The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. - DOI - PubMed
-
- Eguale T., Gebreyes W.A., Asrat D., Alemayehu H., Gunn J.S., Engidawork E. Non-Typhoidal Salmonella Serotypes, Antimicrobial Resistance and Co-Infection with Parasites among Patients with Diarrhea and Other Gastrointestinal Complaints in Addis Ababa, Ethiopia. BMC Infect. Dis. 2015;15:497. doi: 10.1186/s12879-015-1235-y. - DOI - PMC - PubMed
-
- Balasubramanian R., Im J., Lee J.-S., Jeon H.J., Mogeni O.D., Kim J.H., Rakotozandrindrainy R., Baker S., Marks F. The Global Burden and Epidemiology of Invasive Non-Typhoidal Salmonella Infections. Hum. Vaccines Immunother. 2019;15:1421–1426. doi: 10.1080/21645515.2018.1504717. - DOI - PMC - PubMed
-
- Grimont P.A.D., Weill F.-X. Antigenic Formulae of the Salmonella Serovars. Volume 9. WHO Collaborating Centre for Reference and Research on Salmonella; Paris, France: 2007. pp. 1–166.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

