Identification of Potential Treatments for Acute Lymphoblastic Leukemia through Integrated Genomic Network Analysis
- PMID: 36559013
- PMCID: PMC9786277
- DOI: 10.3390/ph15121562
Identification of Potential Treatments for Acute Lymphoblastic Leukemia through Integrated Genomic Network Analysis
Abstract
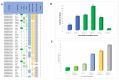
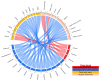
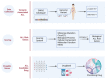
The advancement of high-throughput sequencing and genomic analysis revealed that acute lymphoblastic leukemia (ALL) is a genetically heterogeneous disease. The abundance of such genetic data in ALL can also be utilized to identify potential targets for drug discovery and even drug repurposing. We aimed to determine potential genes for drug development and further guide the identification of candidate drugs repurposed for treating ALL through integrated genomic network analysis. Genetic variants associated with ALL were retrieved from the GWAS Catalog. We further applied a genomic-driven drug repurposing approach based on the six functional annotations to prioritize crucial biological ALL-related genes based on the scoring system. Lastly, we identified the potential drugs in which the mechanisms overlapped with the therapeutic targets and prioritized the candidate drugs using Connectivity Map (CMap) analysis. Forty-two genes were considered biological ALL-risk genes with ARID5B topping the list. Based on potentially druggable genes that we identified, palbociclib, sirolimus, and tacrolimus were under clinical trial for ALL. Additionally, chlorprothixene, sirolimus, dihydroergocristine, papaverine, and tamoxifen are the top five drug repositioning candidates for ALL according to the CMap score with dasatinib as a comparator. In conclusion, this study determines the practicability and the potential of integrated genomic network analysis in driving drug discovery in ALL.
Keywords: acute lymphoblastic leukemia; bioinformatics; drug repurposing; genetic variants; genomic network analysis; leukemia.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
The use of genomic variants to drive drug repurposing for chronic hepatitis B.Biochem Biophys Rep. 2022 Jul 8;31:101307. doi: 10.1016/j.bbrep.2022.101307. eCollection 2022 Sep. Biochem Biophys Rep. 2022. PMID: 35832745 Free PMC article.
-
Integration of genetic variants and gene network for drug repurposing in colorectal cancer.Pharmacol Res. 2020 Nov;161:105203. doi: 10.1016/j.phrs.2020.105203. Epub 2020 Sep 17. Pharmacol Res. 2020. PMID: 32950641
-
Integrated genomic analysis to identify druggable targets for pancreatic cancer.Front Oncol. 2022 Dec 1;12:989077. doi: 10.3389/fonc.2022.989077. eCollection 2022. Front Oncol. 2022. PMID: 36531045 Free PMC article.
-
Integration of genomic variants and bioinformatic-based approach to drive drug repurposing for multiple sclerosis.Biochem Biophys Rep. 2022 Sep 5;32:101337. doi: 10.1016/j.bbrep.2022.101337. eCollection 2022 Dec. Biochem Biophys Rep. 2022. PMID: 36105612 Free PMC article.
-
Turning genome-wide association study findings into opportunities for drug repositioning.Comput Struct Biotechnol J. 2020 Jun 12;18:1639-1650. doi: 10.1016/j.csbj.2020.06.015. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32670504 Free PMC article. Review.
Cited by
-
Nanodrug Delivery Systems for Acute Lymphoblastic Leukemia Therapy.Pharmaceuticals (Basel). 2025 Apr 27;18(5):639. doi: 10.3390/ph18050639. Pharmaceuticals (Basel). 2025. PMID: 40430460 Free PMC article. Review.
-
Integrated genomic network analysis revealed potential of a druggable target for hemorrhoid treatment.Saudi Pharm J. 2023 Dec;31(12):101831. doi: 10.1016/j.jsps.2023.101831. Epub 2023 Oct 20. Saudi Pharm J. 2023. PMID: 37965490 Free PMC article.
-
The landscape of the methodology in drug repurposing using human genomic data: a systematic review.Brief Bioinform. 2024 Jan 22;25(2):bbad527. doi: 10.1093/bib/bbad527. Brief Bioinform. 2024. PMID: 38279645 Free PMC article.
References
-
- Brown P.A., Shah B., Advani A., Aoun P., Boyer M.W., Burke P.W., DeAngelo D.J., Dinner S., Fathi A.T., Gauthier J., et al. Acute Lymphoblastic Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021;19:1079–1109. doi: 10.6004/jnccn.2021.0042. - DOI - PubMed
-
- Jabbour E., Dull J., Yilmaz M., Khoury J.D., Ravandi F., Jain N., Einsele H., Garcia-Manero G., Konopleva M., Short N.J., et al. Outcome of patients with relapsed/refractory acute lymphoblastic leukemia after blinatumomab failure: No change in the level of CD19 expression. Am. J. Hematol. 2018;93:371–374. doi: 10.1002/ajh.24987. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

