Norovirus: An Overview of Virology and Preventative Measures
- PMID: 36560815
- PMCID: PMC9781483
- DOI: 10.3390/v14122811
Norovirus: An Overview of Virology and Preventative Measures
Abstract
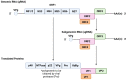
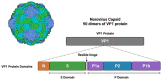
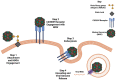
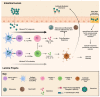
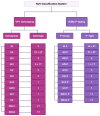
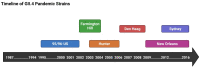
Norovirus (NoV) is an enteric non-enveloped virus which is the leading cause of gastroenteritis across all age groups. It is responsible for around 200,000 deaths annually and outbreaks are common in small communities such as educational and care facilities. 40% of all NoV outbreaks occur in long-term and acute-care facilities, forming the majority of outbreaks. Nosocomial settings set ideal environments for ease of transmission, especially due to the presence of immunocompromised groups. It is estimated to cost global economies around £48 billion a year, making it a global issue. NoV is transmitted via the faecal-oral route and infection with it results in asymptomatic cases or gastrointestinal disease. It has high mutational rates and this allows for new variants to emerge and be more resistant. The classification system available divides NoV into 10 genogroups and 49 genotypes based on whole amino acid sequencing of VP1 capsid protein and partial sequencing of RdRp, respectively. The most predominant genotypes which cause gastroenteritis in humans include GI.1 and GII.4, where GII.4 is responsible for more extreme clinical implications such as hospitalisation. In addition, GII.4 has been responsible for 6 pandemic strains, the last of which is the GII.4 Sydney (2012) variant. In recent years, the successful cultivation of HuNoV was reported in stem cell-derived human intestinal enteroids (HIEs), which promises to assist in giving a deeper understanding of its underlying mechanisms of infection and the development of more personalized control measures. There are no specific control measures against NoV, therefore common practices are used against it such as hand washing. No vaccine is available, but the HIL-214 candidate passed clinical phase 2b and shows promise.
Keywords: classification; control measures; epidemiology; genome; genotype; norovirus; outbreak prevention; pandemics and transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chan M.C., Kwan H.S., Chan P.K. Structure and Genotypes of Noroviruses. Norovirus. 2017;1:51–63.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

