Cyclospora cayetanensis comprises at least 3 species that cause human cyclosporiasis
- PMID: 36560856
- PMCID: PMC10090632
- DOI: 10.1017/S003118202200172X
Cyclospora cayetanensis comprises at least 3 species that cause human cyclosporiasis
Abstract
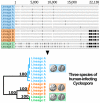
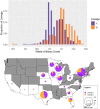
The apicomplexan parasite Cyclospora cayetanensis causes seasonal foodborne outbreaks of the gastrointestinal illness cyclosporiasis. Prior to the coronavirus disease-2019 pandemic, annually reported cases were increasing in the USA, leading the US Centers for Disease Control and Prevention to develop a genotyping tool to complement cyclosporiasis outbreak investigations. Thousands of US isolates and 1 from China (strain CHN_HEN01) were genotyped by Illumina amplicon sequencing, revealing 2 lineages (A and B). The allelic composition of isolates was examined at each locus. Two nuclear loci (CDS3 and 360i2) distinguished lineages A and B. CDS3 had 2 major alleles: 1 almost exclusive to lineage A and the other to lineage B. Six 360i2 alleles were observed – 2 exclusive to lineage A (alleles A1 and A2), 2 to lineage B (B1 and B2) and 1 (B4) was exclusive to CHN_HEN01 which shared allele B3 with lineage B. Examination of heterozygous genotypes revealed that mixtures of A- and B-type 360i2 alleles occurred rarely, suggesting a lack of gene flow between lineages. Phylogenetic analysis of loci from whole-genome shotgun sequences, mitochondrial and apicoplast genomes, revealed that CHN_HEN01 represents a distinct lineage (C). Retrospective examination of epidemiologic data revealed associations between lineage and the geographical distribution of US infections plus strong temporal associations. Given the multiple lines of evidence for speciation within human-infecting Cyclospora, we provide an updated taxonomic description of C. cayetanensis, and describe 2 novel species as aetiological agents of human cyclosporiasis: Cyclospora ashfordi sp. nov. and Cyclospora henanensis sp. nov. (Apicomplexa: Eimeriidae).
Keywords: Cyclospora ashfordi; Cyclospora cayetanensis; Cyclospora henanensis; epidemiology; genotyping; speciation.
Figures





References
-
- Anonymous CfDCaP (1997) Update: outbreaks of cyclosporiasis – United States and Canada, 1997. MMWR. Morbidity and Mortality Weekly Report 46, 521–523. - PubMed
-
- Ashford RW (1979) Occurrence of an undescribed coccidian in man in Papua New Guinea. Annals of Tropical Medicine & Parasitology 73, 497–500. - PubMed
-
- Babiker HA, Ranford-Cartwright LC, Currie D, Charlwood JD, Billingsley P, Teuscher T and Walliker D (1994) Random mating in a natural population of the malaria parasite Plasmodium falciparum. Parasitology 109, 413–421. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

