Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C
- PMID: 36561072
- PMCID: PMC9749929
- DOI: 10.1039/d2md00246a
Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C
Abstract
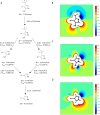
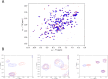
The cellular tumor antigen p53 is a key component in cell cycle control. The mutation Y220C heavily destabilizes the protein thermally but yields a druggable crevice. We have screened the diversity-optimized halogen-enriched fragment library against T-p53C-Y220C with STD-NMR and DSF to identify hits, which we validated by 1H,15N-HSQC NMR. We could identify four hits binding in the Y220C cleft, one hit binding covalently and four hits binding to an uncharacterized binding site. Compound 1151 could be crystallized showing a flip of C220 and thus opening subsite 3. Additionally, 4482 was identified to alkylate cysteines. Data shows that the diversity-optimized HEFLib leads to multiple diverse hits. The identified scaffolds can be used to further optimize interactions with T-p53C-Y220C and increase thermal stability.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures







References
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials
Miscellaneous

