This is a preprint.
Live-attenuated pediatric parainfluenza vaccine expressing 6P-stabilized SARS-CoV-2 spike protein is protective against SARS-CoV-2 variants in hamsters
- PMID: 36561185
- PMCID: PMC9774222
- DOI: 10.1101/2022.12.12.520032
Live-attenuated pediatric parainfluenza vaccine expressing 6P-stabilized SARS-CoV-2 spike protein is protective against SARS-CoV-2 variants in hamsters
Update in
-
Live-attenuated pediatric parainfluenza vaccine expressing 6P-stabilized SARS-CoV-2 spike protein is protective against SARS-CoV-2 variants in hamsters.PLoS Pathog. 2023 Jun 23;19(6):e1011057. doi: 10.1371/journal.ppat.1011057. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37352333 Free PMC article.
Abstract
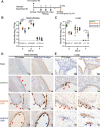
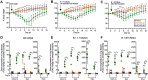
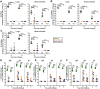
The pediatric live-attenuated bovine/human parainfluenza virus type 3 (B/HPIV3)-vectored vaccine expressing the prefusion-stabilized SARS-CoV-2 spike (S) protein (B/HPIV3/S-2P) was previously evaluated in vitro and in hamsters. To improve its immunogenicity, we generated B/HPIV3/S-6P, expressing S further stabilized with 6 proline mutations (S-6P). Intranasal immunization of hamsters with B/HPIV3/S-6P reproducibly elicited significantly higher serum anti-S IgA/IgG titers than B/HPIV3/S-2P; hamster sera efficiently neutralized variants of concern (VoCs), including Omicron variants. B/HPIV3/S-2P and B/HPIV3/S-6P immunization protected hamsters against weight loss and lung inflammation following SARS-CoV-2 challenge with the vaccine-matched strain WA1/2020 or VoCs B.1.1.7/Alpha or B.1.351/Beta and induced near-sterilizing immunity. Three weeks post-challenge, B/HPIV3/S-2P- and B/HPIV3/S-6P-immunized hamsters exhibited a robust anamnestic serum antibody response with increased neutralizing potency to VoCs, including Omicron sublineages. B/HPIV3/S-6P primed for stronger anamnestic antibody responses after challenge with WA1/2020 than B/HPIV3/S-2P. B/HPIV3/S-6P will be evaluated as an intranasal vaccine to protect infants against both HPIV3 and SARS-CoV-2.
Author summary: SARS-CoV-2 infects and causes disease in all age groups. While injectable SARS-CoV-2 vaccines are effective against severe COVID-19, they do not fully prevent SARS-CoV-2 replication and transmission. This study describes the preclinical comparison in hamsters of B/HPIV3/S-2P and B/HPIV3/S-6P, live-attenuated pediatric vector vaccine candidates expressing the "2P" prefusion stabilized version of the SARS-CoV-2 spike protein, or the further-stabilized "6P" version. B/HPIV3/S-6P induced significantly stronger anti-S serum IgA and IgG responses than B/HPIV3/S-2P. A single intranasal immunization with B/HPIV3/S-6P elicited broad systemic antibody responses in hamsters that efficiently neutralized the vaccine-matched isolate as well as variants of concern, including Omicron. B/HPIV3/S-6P immunization induced near-complete airway protection against the vaccine-matched SARS-CoV-2 isolate as well as two variants. Furthermore, following SARS-CoV-2 challenge, immunized hamsters exhibited strong anamnestic serum antibody responses. Based on these data, B/HPIV3/S-6P will be further evaluated in a phase I study.
Conflict of interest statement
DECLARATION OF INTERESTS
U.J.B., C.L., X.L, and C.LN. are inventors on the provisional patent application number 63/180,534, entitled “Recombinant chimeric bovine/human parainfluenza virus 3 expressing SARS-CoV-2 spike protein and its use”, filed by the United States of America, Department of Health and Human Services.
Figures



Similar articles
-
Live-attenuated pediatric parainfluenza vaccine expressing 6P-stabilized SARS-CoV-2 spike protein is protective against SARS-CoV-2 variants in hamsters.PLoS Pathog. 2023 Jun 23;19(6):e1011057. doi: 10.1371/journal.ppat.1011057. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37352333 Free PMC article.
-
Intranasal parainfluenza virus-vectored vaccine expressing SARS-CoV-2 spike protein of Delta or Omicron B.1.1.529 induces mucosal and systemic immunity and protects hamsters against homologous and heterologous challenge.PLoS Pathog. 2025 Apr 21;21(4):e1012585. doi: 10.1371/journal.ppat.1012585. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40258004 Free PMC article.
-
A single intranasal dose of a live-attenuated parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in hamsters.Proc Natl Acad Sci U S A. 2021 Dec 14;118(50):e2109744118. doi: 10.1073/pnas.2109744118. Proc Natl Acad Sci U S A. 2021. PMID: 34876520 Free PMC article.
-
Intranasal parainfluenza virus-vectored vaccine expressing SARS-CoV-2 spike protein of Delta or Omicron B.1.1.529 induces mucosal and systemic immunity and protects hamsters against homologous and heterologous challenge.bioRxiv [Preprint]. 2024 Sep 13:2024.09.12.612598. doi: 10.1101/2024.09.12.612598. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Apr 21;21(4):e1012585. doi: 10.1371/journal.ppat.1012585. PMID: 39372768 Free PMC article. Updated. Preprint.
-
Intranasal pediatric parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in monkeys.Cell. 2022 Dec 8;185(25):4811-4825.e17. doi: 10.1016/j.cell.2022.11.006. Epub 2022 Nov 10. Cell. 2022. PMID: 36423629 Free PMC article.
Cited by
-
Versatility of reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) from diagnosis of early pathological infection to mutation detection in organisms.Mol Biol Rep. 2024 Jan 25;51(1):211. doi: 10.1007/s11033-023-09110-z. Mol Biol Rep. 2024. PMID: 38270670 Review.
References
-
- Martin B, DeWitt PE, Russell S, Anand A, Bradwell KR, Bremer C, et al. Characteristics, Outcomes, and Severity Risk Factors Associated With SARS-CoV-2 Infection Among Children in the US National COVID Cohort Collaborative. JAMA Netw Open. 2022;5(2):e2143151. Epub 20220201. doi: 10.1001/jamanetworkopen.2021.43151. - DOI - PMC - PubMed
-
- Zachariah P, Johnson CL, Halabi KC, Ahn D, Sen AI, Fischer A, et al. Epidemiology, Clinical Features, and Disease Severity in Patients With Coronavirus Disease 2019 (COVID-19) in a Children’s Hospital in New York City, New York. JAMA Pediatr. 2020;174(10):e202430. Epub 20201005. doi: 10.1001/jamapediatrics.2020.2430. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
