A non-destructive coconut fruit and seed traits extraction method based on Micro-CT and deeplabV3+ model
- PMID: 36561444
- PMCID: PMC9763456
- DOI: 10.3389/fpls.2022.1069849
A non-destructive coconut fruit and seed traits extraction method based on Micro-CT and deeplabV3+ model
Abstract
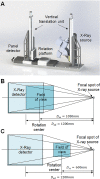
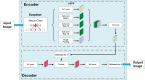
With the completion of the coconut gene map and the gradual improvement of related molecular biology tools, molecular marker-assisted breeding of coconut has become the next focus of coconut breeding, and accurate coconut phenotypic traits measurement will provide technical support for screening and identifying the correspondence between genotype and phenotype. A Micro-CT system was developed to measure coconut fruits and seeds automatically and nondestructively to acquire the 3D model and phenotyping traits. A deeplabv3+ model with an Xception backbone was used to segment the sectional image of coconut fruits and seeds automatically. Compared with the structural-light system measurement, the mean absolute percentage error of the fruit volume and surface area measurements by the Micro-CT system was 1.87% and 2.24%, respectively, and the squares of the correlation coefficients were 0.977 and 0.964, respectively. In addition, compared with the manual measurements, the mean absolute percentage error of the automatic copra weight and total biomass measurements was 8.85% and 25.19%, respectively, and the adjusted squares of the correlation coefficients were 0.922 and 0.721, respectively. The Micro-CT system can nondestructively obtain up to 21 agronomic traits and 57 digital traits precisely.
Keywords: Micro-CT; coconut phenotypic traits; deep learning; non-destructive; plant phenomics.
Copyright © 2022 Yu, Liu, Yang, Wu, Wang, He, Chen and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Advanced deep learning models for phenotypic trait extraction and cultivar classification in lychee using photon-counting micro-CT imaging.Front Plant Sci. 2024 Feb 29;15:1358360. doi: 10.3389/fpls.2024.1358360. eCollection 2024. Front Plant Sci. 2024. PMID: 38486848 Free PMC article.
-
A deep learning-integrated micro-CT image analysis pipeline for quantifying rice lodging resistance-related traits.Plant Commun. 2021 Jan 29;2(2):100165. doi: 10.1016/j.xplc.2021.100165. eCollection 2021 Mar 8. Plant Commun. 2021. PMID: 33898978 Free PMC article.
-
High-Throughput Phenotyping of Morphological Seed and Fruit Characteristics Using X-Ray Computed Tomography.Front Plant Sci. 2020 Nov 12;11:601475. doi: 10.3389/fpls.2020.601475. eCollection 2020. Front Plant Sci. 2020. PMID: 33281857 Free PMC article.
-
Recent Advances in Sugarcane Genomics, Physiology, and Phenomics for Superior Agronomic Traits.Front Genet. 2022 Aug 3;13:854936. doi: 10.3389/fgene.2022.854936. eCollection 2022. Front Genet. 2022. PMID: 35991570 Free PMC article. Review.
-
Fruit Biology of Coconut (Cocos nucifera L.).Plants (Basel). 2022 Nov 29;11(23):3293. doi: 10.3390/plants11233293. Plants (Basel). 2022. PMID: 36501334 Free PMC article. Review.
Cited by
-
Research on the evolutionary history of the morphological structure of cotton seeds: a new perspective based on high-resolution micro-CT technology.Front Plant Sci. 2023 Oct 13;14:1219476. doi: 10.3389/fpls.2023.1219476. eCollection 2023. Front Plant Sci. 2023. PMID: 37900733 Free PMC article.
-
CT image segmentation of foxtail millet seeds based on semantic segmentation model VGG16-UNet.Plant Methods. 2024 Nov 7;20(1):169. doi: 10.1186/s13007-024-01288-y. Plant Methods. 2024. PMID: 39511624 Free PMC article.
-
Preliminary study on the association between lignan metabolites and CT non-destructive testing of coconut fruit at different developmental stages.PeerJ. 2024 Sep 26;12:e18049. doi: 10.7717/peerj.18049. eCollection 2024. PeerJ. 2024. PMID: 39346073 Free PMC article.
-
Advanced deep learning models for phenotypic trait extraction and cultivar classification in lychee using photon-counting micro-CT imaging.Front Plant Sci. 2024 Feb 29;15:1358360. doi: 10.3389/fpls.2024.1358360. eCollection 2024. Front Plant Sci. 2024. PMID: 38486848 Free PMC article.
-
An improved Deeplab V3+ network based coconut CT image segmentation method.Front Plant Sci. 2023 Dec 8;14:1139666. doi: 10.3389/fpls.2023.1139666. eCollection 2023. Front Plant Sci. 2023. PMID: 38148865 Free PMC article.
References
-
- Abadi Martín, Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., et al. . (2015) TensorFlow: Large-scale machine learning on heterogeneous systems. Available at: https://www.tensorflow.org/.
-
- Beevi S. N., Mohan P., Paul A., Mathew T. B. (2006). Germination and seedling characters in coconut (Cocos nucifera l.) as affected by eriophyid mite (Aceria guerreronis keifer) infestation. J. Trop. Agric. 44 (1-2), 76–78.
-
- Caladcad J. A., Cabahug S., Catamco M. R., Villaceran P. E., Cosgafa L., Cabizares K. N., et al. . (2020). Determining Philippine coconut maturity level using machine learning algorithms based on acoustic signal. Comput. Electron. Agric. 172, 105327. doi: 10.1016/j.compag.2020.105327 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

