Neutrophils and macrophages drive TNF-induced lethality via TRIF/CD14-mediated responses
- PMID: 36563168
- PMCID: PMC10021564
- DOI: 10.1126/sciimmunol.add0665
Neutrophils and macrophages drive TNF-induced lethality via TRIF/CD14-mediated responses
Abstract
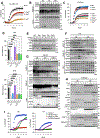
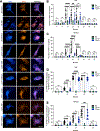
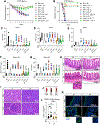
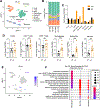
TNF mediates a variety of biological processes including cellular proliferation, inflammatory responses, and cell death and is therefore associated with numerous pathologies including autoinflammatory diseases and septic shock. The inflammatory and cell death responses to TNF have been studied extensively downstream of TNF-R1 and are believed to rely on the formation of proinflammatory complex I and prodeath complex II, respectively. We recently identified a similar multimeric complex downstream of TLR4, termed the TRIFosome, that regulates inflammation and cell death in response to LPS or Yersinia pseudotuberculosis. We present evidence of a role for the TRIFosome downstream of TNF-R1, independent of TLR3 or TLR4 engagement. Specifically, TNF-induced cell death and inflammation in murine macrophages were driven by the TLR4 adaptor TRIF and the LPS co-receptor CD14, highlighting an important role for these proteins beyond TLR-mediated immune responses. Via immunoprecipitation and visualization of TRIF-specific puncta, we demonstrated TRIF- and CD14-dependent formation of prodeath and proinflammatory complexes in response to TNF. Extending these findings, in a murine TNF-induced sepsis model, TRIF and CD14 deficiency decreased systemic inflammation, reduced organ pathology, and improved survival. The outcome of TRIF activation was cell specific, because TNF-induced lethality was mediated by neutrophils and macrophages responding to TNF in a TRIF-dependent manner. Our findings suggest that in addition to their crucial role in TNF production, myeloid cells are central to TNF toxicity and position TRIF and CD14 as universal components of receptor-mediated immune responses.
Conflict of interest statement
Figures


References
-
- Mcdermott MF, Aksentijevich I, Mcdermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, Mccarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo A, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Amos CI, Mulley J, Quane KA, Molloy MG, Ranki A, Powell RJ, Hitman GA, Shea JJO, Kastner DL, Germline Mutations in the Extracellular Domains of the 55 kDa TNF Receptor, TNFR1, Define a Family of Dominantly Inherited Autoinflammatory Syndromes. Cell 97, 133–144 (1999). - PubMed
-
- Garcia-carbonell R, Wong J, Kim JY, Close LA, Boland BS, Wong TL, Harris PA, Ho SB, Das S, Ernst PB, Sasik R, Sandborn WJ, Bertin J, Gough PJ, Chang JT, Kelliher MA, Boone D, Guma M, Karin M, Elevated A20 promotes TNF-induced and RIPK1- dependent intestinal epithelial cell death. PNAS 115, 9192–9200 (2018). - PMC - PubMed
-
- Del Valle DM, Kim-schulze S, Huang H, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M, Gnjatic S, An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

