Measurement of Adenovirus-Based Vector Heterogeneity
- PMID: 36563855
- PMCID: PMC9767660
- DOI: 10.1016/j.xphs.2022.12.012
Measurement of Adenovirus-Based Vector Heterogeneity
Abstract
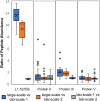
Adenovirus vectors have become an important class of vaccines with the recent approval of Ebola and COVID-19 products. In-process quality attribute data collected during Adenovirus vector manufacturing has focused on particle concentration and infectivity ratios (based on viral genome: cell-based infectivity), and data suggest only a fraction of viral particles present in the final vaccine product are efficacious. To better understand this product heterogeneity, lab-scale preparations of two Adenovirus viral vectors, (Chimpanzee adenovirus (ChAdOx1) and Human adenovirus Type 5 (Ad5), were studied using transmission electron microscopy (TEM). Different adenovirus morphologies were characterized, and the proportion of empty and full viral particles were quantified. These proportions showed a qualitative correlation with the sample's infectivity values. Liquid chromatography-mass spectrometry (LC-MS) peptide mapping was used to identify key adenovirus proteins involved in viral maturation. Using peptide abundance analysis, a ∼5-fold change in L1 52/55k abundance was observed between low-(empty) and high-density (full) fractions taken from CsCl ultracentrifugation preparations of ChAdOx1 virus. The L1 52/55k viral protein is associated with DNA packaging and is cleaved during viral maturation, so it may be a marker for infective particles. TEM and LC-MS peptide mapping are promising higher-resolution analytical characterization tools to help differentiate between relative proportions of empty, non-infectious, and infectious viral particles as part of Adenovirus vector in-process monitoring, and these results are an encouraging initial step to better differentiate between the different product-related impurities.
Keywords: Adenovirus-based vaccine; Analytical characterization; Critical quality attributes; In-process testing; Mass spectrometry; Transmission electron microscopy.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interests The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Barnes LF, Draper BE, Jarrold MF. Analysis of recombinant adenovirus vectors by ion trap charge detection mass spectrometry: accurate molecular weight measurements beyond 150 MDa. Anal Chem. 2022;94(3):1543–1551. - PubMed
-
- Evans RK, Nawrocki DK, Isopi LA, et al. Development of stable liquid formulations for adenovirus-based vaccines. J Pharm Sci. 2004;93(10):2458–2475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

