Elucidation of ubiquitin-conjugating enzymes that interact with RBR-type ubiquitin ligases using a liquid-liquid phase separation-based method
- PMID: 36563856
- PMCID: PMC9860496
- DOI: 10.1016/j.jbc.2022.102822
Elucidation of ubiquitin-conjugating enzymes that interact with RBR-type ubiquitin ligases using a liquid-liquid phase separation-based method
Abstract
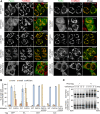
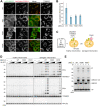
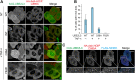
RING-between RING (RBR)-type ubiquitin (Ub) ligases (E3s) such as Parkin receive Ub from Ub-conjugating enzymes (E2s) in response to ligase activation. However, the specific E2s that transfer Ub to each RBR-type ligase are largely unknown because of insufficient methods for monitoring their interaction. To address this problem, we have developed a method that detects intracellular interactions between E2s and activated Parkin. Fluorescent homotetramer Azami-Green fused with E2 and oligomeric Ash (Assembly helper) fused with Parkin form a liquid-liquid phase separation (LLPS) in cells only when E2 and Parkin interact. Using this method, we identified multiple E2s interacting with activated Parkin on damaged mitochondria during mitophagy. Combined with in vitro ubiquitination assays and bioinformatics, these findings revealed an underlying consensus sequence for E2 interactions with activated Parkin. Application of this method to other RBR-type E3s including HOIP, HHARI, and TRIAD1 revealed that HOIP forms an LLPS with its substrate NEMO in response to a proinflammatory cytokine and that HHARI and TRIAD1 form a cytosolic LLPS independent of Ub-like protein NEDD8. Since an E2-E3 interaction is a prerequisite for RBR-type E3 activation and subsequent substrate ubiquitination, the method we have established here can be an in-cell tool to elucidate the potentially novel mechanisms involved in RBR-type E3s.
Keywords: PINK1; Parkin; RBR-type E3; mitochondria; mitophagy; ubiquitin.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Trempe J.F., Sauve V., Grenier K., Seirafi M., Tang M.Y., Menade M., et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

