PCNA regulates primary metabolism by scaffolding metabolic enzymes
- PMID: 36564470
- PMCID: PMC9937922
- DOI: 10.1038/s41388-022-02579-1
PCNA regulates primary metabolism by scaffolding metabolic enzymes
Abstract
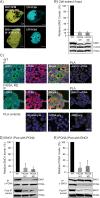
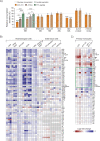
The essential roles of proliferating cell nuclear antigen (PCNA) as a scaffold protein in DNA replication and repair are well established, while its cytosolic roles are less explored. Two metabolic enzymes, alpha-enolase (ENO1) and 6-phosphogluconate dehydrogenase (6PGD), both contain PCNA interacting motifs. Mutation of the PCNA interacting motif APIM in ENO1 (F423A) impaired its binding to PCNA and resulted in reduced cellular levels of ENO1 protein, reduced growth rate, reduced glucose consumption, and reduced activation of AKT. Metabolome and signalome analysis reveal large consequences of impairing the direct interaction between PCNA and ENO1. Metabolites above ENO1 in glycolysis accumulated while lower glycolytic and TCA cycle metabolite pools decreased in the APIM-mutated cells; however, their overall energetic status were similar to parental cells. Treating haematological cancer cells or activated primary monocytes with a PCNA targeting peptide drug containing APIM (ATX-101) also lead to a metabolic shift characterized by reduced glycolytic rate. In addition, we show that ATX-101 treatments reduced the ENO1 - PCNA interaction, the ENO1, GAPDH and 6PGD protein levels, as well as the 6PGD activity. Here we report for the first time that PCNA acts as a scaffold for metabolic enzymes, and thereby act as a direct regulator of primary metabolism.
© 2022. The Author(s).
Conflict of interest statement
APIM Therapeutics is a spin-off company of the Norwegian University of Science and Technology, NTNU, developing APIM-peptides for use in cancer therapy. The lead APIM-peptide ATX-101 is currently in Phase II. Professor Marit Otterlei is founder, minority shareholder and part time CSO of this company. The authors have no additional competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

