Short-chain fatty acid-mediated epigenetic modulation of inflammatory T cells in vitro
- PMID: 36566262
- PMCID: PMC10695156
- DOI: 10.1007/s13346-022-01284-6
Short-chain fatty acid-mediated epigenetic modulation of inflammatory T cells in vitro
Abstract
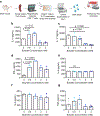
Short-chain fatty acids (SCFAs) are major metabolic products of indigestible polysaccharides in the gut and mediate the function of immune cells to facilitate homeostasis. The immunomodulatory effect of SCFAs has been attributed, at least in part, to the epigenetic modulation of immune cells through the inhibition the nucleus-resident enzyme histone deacetylase (HDAC). Among the downstream effects, SCFAs enhance regulatory T cells (Treg) over inflammatory T helper (Th) cells, including Th17 cells, which can be pathogenic. Here, we characterize the potential of two common SCFAs-butyrate and pentanoate-in modulating differentiation of T cells in vitro. We show that butyrate but not pentanoate exerts a concentration-dependent effect on Treg and Th17 differentiation. Increasing the concentration of butyrate suppresses the Th17-associated RORγtt and IL-17 and increases the expression of Treg-associated FoxP3. To effectively deliver butyrate, encapsulation of butyrate in a liposomal carrier, termed BLIPs, reduced cytotoxicity while maintaining the immunomodulatory effect on T cells. Consistent with these results, butyrate and BLIPs inhibit HDAC and promote a unique chromatin landscape in T cells under conditions that otherwise promote conversion into a pro-inflammatory phenotype. Motif enrichment analysis revealed that butyrate and BLIP-mediated suppression of Th17-associated chromatin accessibility corresponded with a marked decrease in bZIP family transcription factor binding sites. These results support the utility and further evaluation of BLIPs as an immunomodulatory agent for autoimmune disorders that are characterized by chronic inflammation and pathogenic inflammatory T cells.
Keywords: Epigenetic modulation; Immunomodulation; Short-chain fatty acids; T cells.
© 2022. Controlled Release Society.
Conflict of interest statement
Competing interests:
The authors have no relevant financial or non-financial interests to disclose
Figures




References
-
- Paust S. & Cantor H. Regulatory T cells and autoimmune disease. Immunol Rev 204, 195–207 (2005). - PubMed
-
- Ohl K. & Tenbrock K. Regulatory T cells in systemic lupus erythematosus. Eur J Immunol 45, 344–355 (2015). - PubMed
-
- Littman DR & Rudensky AY Th17 and Regulatory T Cells in Mediating and Restraining Inflammation. Cell 140, 845–858 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

