Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
- PMID: 36567772
- PMCID: PMC9775220
- DOI: 10.1021/acscatal.2c00336
Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
Abstract
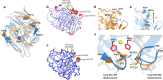
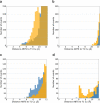
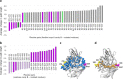
Laccases are in increasing demand as innovative solutions in the biorefinery fields. Here, we combine mutagenesis with structural, kinetic, and in silico analyses to characterize the molecular features that cause the evolution of a hyperthermostable metallo-oxidase from the multicopper oxidase family into a laccase (k cat 273 s-1 for a bulky aromatic substrate). We show that six mutations scattered across the enzyme collectively modulate dynamics to improve the binding and catalysis of a bulky aromatic substrate. The replacement of residues during the early stages of evolution is a stepping stone for altering the shape and size of substrate-binding sites. Binding sites are then fine-tuned through high-order epistasis interactions by inserting distal mutations during later stages of evolution. Allosterically coupled, long-range dynamic networks favor catalytically competent conformational states that are more suitable for recognizing and stabilizing the aromatic substrate. This work provides mechanistic insight into enzymatic and evolutionary molecular mechanisms and spots the importance of iterative experimental and computational analyses to understand local-to-global changes.
© 2022 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous
