A differential process mining analysis of COVID-19 management for cancer patients
- PMID: 36568192
- PMCID: PMC9768429
- DOI: 10.3389/fonc.2022.1043675
A differential process mining analysis of COVID-19 management for cancer patients
Erratum in
-
Erratum: A differential process mining analysis of COVID-19 management for cancer patients.Front Oncol. 2023 Mar 8;13:1173233. doi: 10.3389/fonc.2023.1173233. eCollection 2023. Front Oncol. 2023. PMID: 36969046 Free PMC article.
Abstract
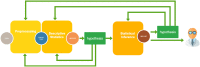
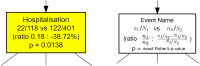
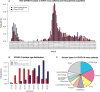
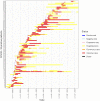
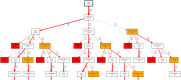
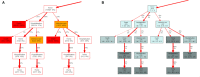
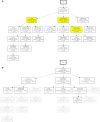
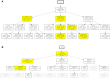
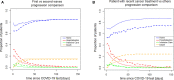
During the acute phase of the COVID-19 pandemic, hospitals faced a challenge to manage patients, especially those with other comorbidities and medical needs, such as cancer patients. Here, we use Process Mining to analyze real-world therapeutic pathways in a cohort of 1182 cancer patients of the Lausanne University Hospital following COVID-19 infection. The algorithm builds trees representing sequences of coarse-grained events such as Home, Hospitalization, Intensive Care and Death. The same trees can also show probability of death or time-to-event statistics in each node. We introduce a new tool, called Differential Process Mining, which enables comparison of two patient strata in each node of the tree, in terms of hits and death rate, together with a statistical significance test. We thus compare management of COVID-19 patients with an active cancer in the first vs. second COVID-19 waves to quantify hospital adaptation to the pandemic. We also compare patients having undergone systemic therapy within 1 year to the rest of the cohort to understand the impact of an active cancer and/or its treatment on COVID-19 outcome. This study demonstrates the value of Process Mining to analyze complex event-based real-world data and generate hypotheses on hospital resource management or on clinical patient care.
Keywords: COVID-19; clinical pathways; oncology; process analysis; process mining.
Copyright © 2022 Cuendet, Gatta, Wicky, Gerard, Dalla-Vale, Tavazzi, Michielin, Delyon, Ferahta, Cesbron, Lofek, Huber, Jankovic, Demicheli, Bouchaab, Digklia, Obeid, Peters, Eicher, Pradervand and Michielin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Drew DA, Nguyen LH, Ma W, Lo CH, Joshi AD, Sikavi D, et al. . Abstract s09-01: Cancer and race: Two important risk factors for covid-19 incidence as captured by the covid symptom study real-time epidemiology tool. Clin Cancer Res (2020) 26:S09–01–S09–01. doi: 10.1158/1557-3265.COVID-19-S09-01 - DOI
-
- Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, et al. . Patients with cancer appear more vulnerable to SARS-CoV-2: A multicenter study during the COVID-19 outbreak. Cancer Discovery (2020) 10:783–91. doi: 10.1158/2159-8290.CD-20-0422 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

