Identification of Signature Genes in the PD-1 Relative Gastric Cancer Using a Combined Analysis of Gene Expression and Methylation Data
- PMID: 36568638
- PMCID: PMC9780002
- DOI: 10.1155/2022/4994815
Identification of Signature Genes in the PD-1 Relative Gastric Cancer Using a Combined Analysis of Gene Expression and Methylation Data
Abstract
Background: The morbidity and mortality rates for gastric cancer (GC) rank second among all cancers, indicating the serious threat it poses to human health, as well as human life. This study aims to identify the pathways and genes as well as investigate the molecular mechanisms of tumor-related genes in gastric cancer (GC).
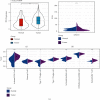
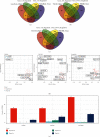
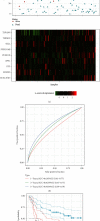
Method: We compared differentially expressed genes (DEGs) and differentially methylated genes (DMGs) in gastric cancer and normal tissue samples using The Cancer Genome Atlas (TCGA) data. The Kyoto Encyclopedia of Gene and Genome (KEGG) and the Gene Ontology (GO) enrichment analysis' pathway annotations were conducted on DMGs and DEGs using a clusterProfiler R package to identify the important functions, as well as the biological processes and pathways involved. The intersection of the two was chosen and defined as differentially methylated and expressed genes (DMEGs). For DMEGs, we used the principal component analysis (PCA) to differentiate gastric cancer from adjacent samples. The linear discriminant analysis method was applied to categorize the samples using DMEGs methylation data and DMEGs expression profiles data and was validated using the leave-one-out cross-validation (LOOCV) method. We plotted the ROC curve for the classification and calculated the AUC (area under the ROC curve) value for a more intuitive view of the classification effect. We also used the NetworkAnalyst 3.0 tool to analyze DMEGs, using DrugBank to acquire information on protein-drug interactions and generate a network map of gene-drug interactions.
Results: We identified a total of 971 DMGs in 188 PD-1 negative and 187 PD-1 positive gastric cancer samples obtained from TCGA. The KEGG and GO enrichment analysis showed the involvement of the regulation of ion transmembrane transport, collagen-containing extracellular matrix, cell-cell junction, and peptidase regulator activity. We simultaneously obtained 1,189 DEGs, out of which 986 were downregulated, while 203 were upregulated in tumors. The enriched analysis of the GO's and KEGG's pathways indicated that the most significant pathways included an intestinal immune network for IgA production, Staphylococcus aureus infection, cytokine-cytokine receptor interaction, and viral protein interaction with cytokine and cytokine receptor, which have previously been linked with gastric cancer. The compound DB01830 can bind well to the active site of the LCK protein and shows good stability, thus making it a potential inhibitor of the LCK protein. To observe the relationship between DMEGs' expression and prognosis, we observed 10 genes, among which were TRIM29, TSPAN8, EOMES, PPP1R16B, SELL, PCED1B, IYD, JPH1, CEACAM5, and RP11-44K6.2. Their high expressions were related to high risks. Besides, those genes were validated in different internal and external validation sets.
Conclusion: These results may provide potential molecular biological therapy for PD-1 negative gastric cancer.
Copyright © 2022 Han Yu et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures







References
-
- Katai H., Ishikawa T., Akazawa K., et al. Five-year survival analysis of surgically resected gastric cancer cases in Japan: a retrospective analysis of more than 100, 000 patients from the nationwide registry of the Japanese Gastric Cancer Association (2001-2007) Gastric Cancer . 2018;21(1):144–154. doi: 10.1007/s10120-017-0716-7. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

