Targets of histone H3 lysine 9 methyltransferases
- PMID: 36568972
- PMCID: PMC9768651
- DOI: 10.3389/fcell.2022.1026406
Targets of histone H3 lysine 9 methyltransferases
Abstract
Histone H3 lysine 9 di- and trimethylation are well-established marks of constitutively silenced heterochromatin domains found at repetitive DNA elements including pericentromeres, telomeres, and transposons. Loss of heterochromatin at these sites causes genomic instability in the form of aberrant DNA repair, chromosome segregation defects, replication stress, and transposition. H3K9 di- and trimethylation also regulate cell type-specific gene expression during development and form a barrier to cellular reprogramming. However, the role of H3K9 methyltransferases extends beyond histone methylation. There is a growing list of non-histone targets of H3K9 methyltransferases including transcription factors, steroid hormone receptors, histone modifying enzymes, and other chromatin regulatory proteins. Additionally, two classes of H3K9 methyltransferases modulate their own function through automethylation. Here we summarize the structure and function of mammalian H3K9 methyltransferases, their roles in genome regulation and constitutive heterochromatin, as well as the current repertoire of non-histone methylation targets including cases of automethylation.
Keywords: chromatin; heterochromatin; histone; methylation; methyltransferase.
Copyright © 2022 Levinsky, McEdwards, Sethna and Currie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
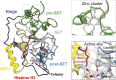
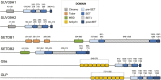

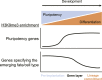
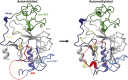
References
Publication types
LinkOut - more resources
Full Text Sources

