Longitudinal serum proteomics analyses identify unique and overlapping host response pathways in Lyme disease and West Nile virus infection
- PMID: 36569838
- PMCID: PMC9784464
- DOI: 10.3389/fimmu.2022.1012824
Longitudinal serum proteomics analyses identify unique and overlapping host response pathways in Lyme disease and West Nile virus infection
Abstract
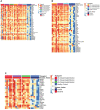
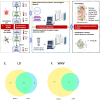
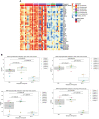
Advancement in proteomics methods for interrogating biological samples has helped identify disease biomarkers for early diagnostics and unravel underlying molecular mechanisms of disease. Herein, we examined the serum proteomes of 23 study participants presenting with one of two common arthropod-borne infections: Lyme disease (LD), an extracellular bacterial infection or West Nile virus infection (WNV), an intracellular viral infection. The LC/MS based serum proteomes of samples collected at the time of diagnosis and during convalescence were assessed using a depletion-based high-throughput shotgun proteomics (dHSP) pipeline as well as a non-depleting blotting-based low-throughput platform (MStern). The LC/MS integrated analyses identified host proteome responses in the acute and recovery phases shared by LD and WNV infections, as well as differentially abundant proteins that were unique to each infection. Notably, we also detected proteins that distinguished localized from disseminated LD and asymptomatic from symptomatic WNV infection. The proteins detected in both diseases with the dHSP pipeline identified unique and overlapping proteins detected with the non-depleting MStern platform, supporting the utility of both detection methods. Machine learning confirmed the use of the serum proteome to distinguish the infection from healthy control sera but could not develop discriminatory models between LD and WNV at current sample numbers. Our study is the first to compare the serum proteomes in two arthropod-borne infections and highlights the similarities in host responses even though the pathogens and the vectors themselves are different.
Keywords: Lyme disease; West Nile virus; acute-phase response; asymptomatic infection; immune system; localized and disseminated stage; longitudinal analysis; serum proteomics.
Copyright © 2022 Boada, Fatou, Belperron, Sigdel, Smolen, Wurie, Levy, Ronca, Murray, Liberto, Rashmi, Kerwin, Montgomery, Bockenstedt, Steen and Sarwal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bennike TB, Bellin MD, Xuan Y, Stensballe A, Moller FT, Beilman GJ, et al. . A cost-effective high-throughput plasma and serum proteomics workflow enables mapping of the molecular impact of total pancreatectomy with islet autotransplantation. J Proteome Res (2018) 17(5):1983–92. doi: 10.1021/acs.jproteome.8b00111 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

