Transcriptional changes in human palate and skin healing
- PMID: 36571451
- PMCID: PMC10006330
- DOI: 10.1111/wrr.13068
Transcriptional changes in human palate and skin healing
Abstract
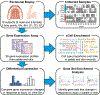
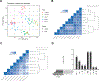
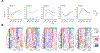
Most human tissue injuries lead to the formation of a fibrous scar and result in the loss of functional tissue. One adult tissue that exhibits a more regenerative response to injury with minimal scarring is the oral mucosa. We generated a microarray gene expression dataset to examine the response to injury in human palate and skin excisional biopsies spanning the first 7 days after wounding. Differential expression analyses were performed in each tissue to identify genes overexpressed or underexpressed over time when compared to baseline unwounded tissue gene expression levels. To attribute biological processes of interest to these gene expression changes, gene set enrichment analysis was used to identify core gene sets that are enriched over the time-course of the wound healing process with respect to unwounded tissue. This analysis identified gene sets uniquely enriched in either palate or skin wounds and gene sets that are enriched in both tissues in at least one time point after injury. Finally, a cell type enrichment analysis was performed to better understand the cell type distribution in these tissues and how it changes over the time course of wound healing. This work provides a source of human wound gene expression data that includes two tissue types with distinct regenerative and scarring phenotypes.
Keywords: genomics; oral mucosa; palate; skin; wound healing.
© 2022 The Authors. Wound Repair and Regeneration published by Wiley Periodicals LLC on behalf of The Wound Healing Society.
Conflict of interest statement
Conflict of Interest
The authors declare no conflicts of interest.
Figures





References
-
- Szpaderska AM, Zuckerman JD, DiPietro LA. Differential injury responses in oral mucosal and cutaneous wounds. J Dent Res 2003;82(8):621–6. - PubMed
-
- Szpaderska AM, Walsh CG, Steinberg MJ, DiPietro LA. Distinct patterns of angiogenesis in oral and skin wounds. J Dent Res 2005;84(4):309–14. - PubMed
-
- Warburton G, Nares S, Angelov N, Brahim JS, Dionne RA, Wahl SM. Transcriptional events in a clinical model of oral mucosal tissue injury and repair. Wound Repair Regen 2005;13(1):19–26. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

