Functional and miRNA regulatory characteristics of INSIG genes highlight the key role of lipid synthesis in the liver of chicken (Gallus gallus)
- PMID: 36571872
- PMCID: PMC9800209
- DOI: 10.1016/j.psj.2022.102380
Functional and miRNA regulatory characteristics of INSIG genes highlight the key role of lipid synthesis in the liver of chicken (Gallus gallus)
Abstract
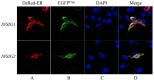
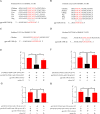
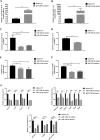
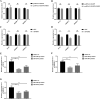
The insulin-induced genes (INSIG1 and INSIG2) have been demonstrated to play a vital role in regulating lipid metabolism in mammals, however the function and regulation mechanism of them remains unknown in poultry. In this study, firstly the phylogenetic trees of INSIGs among various species were constructed and their subcellular locations were mapped in chicken LMH. Then the spatiotemporal expression profiles, over-expression and knockdown assays of chicken INSIGs were conducted. Furthermore, conservation of potential miRNA binding sites in INSIGs among species were analyzed, and the miRNA biological function and regulatory role were verified. The results showed that chicken INSIGs located in cellular endoplasmic reticulum, and were originated from the common ancestors of their mammalian counterparts. The INSIGs were widely expressed in all detected tissues, and their expression levels in the liver of chicken at 30 wk were significantly higher than that at 20 wk (P < 0.01). Over-expression of INSIGs led no significant increase in mRNA abundance of lipid metabolism-related genes and the contents of triacylglycerol (TG) and cholesterol (TC) in LMH cells. Knockdown of INSIG1 led to the decreased expressions of ACSL1, MTTP-L, ApoB, ApoVLDLII genes and TG, TC contents (P < 0.05). Knockdown of INSIG2 could significantly decrease the contents of TG and TC, and expressions of key genes related to the lipid metabolism (P < 0.05). Moreover, INSIG1 was directly targeted by both miR-130b-3p and miR-218-5p, and INSIG2 was directly targeted by miR-130b-3p. MiR-130b-3p mimic and miR-218-5p mimic treatment could significant decrease the mRNA and protein levels of INSIGs, mRNA levels of genes related to lipid metabolism, and the contents of TG and TC in LMH cells. The inhibition of miR-130b-3p and miR-218-5p on TG and TC contents could be restored by the overexpression of INSIGs, respectively. No significant alteration in expressions of sterol regulatory element binding protein (SREBPs) and SREBP cleavage-activating protein (SCAP) were observed when INSIGs were over-expressed. SCAP was down-regulated when INSIG1 was knocked down, while SREBP1 was down-regulated when INSIG2 was knocked down. Taken together, these results highlight the role of INSIG1 and INSIG2 in lipid metabolism and their regulatory mechanism in chicken.
Keywords: INSIG; chicken; lipid metabolism; liver; miRNA.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Insulin-induced gene 1 and 2 isoforms synergistically regulate triacylglycerol accumulation, lipid droplet formation, and lipogenic gene expression in goat mammary epithelial cells.J Dairy Sci. 2019 Feb;102(2):1736-1746. doi: 10.3168/jds.2018-15492. Epub 2018 Dec 26. J Dairy Sci. 2019. PMID: 30594361
-
Regulation of INSIG2 by microRNA-96.Anim Cells Syst (Seoul). 2017 Jun 13;21(4):263-268. doi: 10.1080/19768354.2017.1336483. eCollection 2017. Anim Cells Syst (Seoul). 2017. PMID: 30460077 Free PMC article.
-
miR-19b-3p regulated by estrogen controls lipid synthesis through targeting MSMO1 and ELOVL5 in LMH cells.Poult Sci. 2024 Jan;103(1):103200. doi: 10.1016/j.psj.2023.103200. Epub 2023 Oct 15. Poult Sci. 2024. PMID: 37939591 Free PMC article.
-
Insulin-induced gene: a new regulator in lipid metabolism.Peptides. 2010 Nov;31(11):2145-50. doi: 10.1016/j.peptides.2010.07.020. Epub 2010 Sep 15. Peptides. 2010. PMID: 20817058 Review.
-
Dual functions of Insig proteins in cholesterol homeostasis.Lipids Health Dis. 2012 Dec 18;11:173. doi: 10.1186/1476-511X-11-173. Lipids Health Dis. 2012. PMID: 23249523 Free PMC article. Review.
References
-
- Desvergne B., Michalik L., Wahli W. Transcriptional regulation of metabolism. Physiol. Rev. 2006;86:465–514. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

