Identification of RT-qPCR reference genes suitable for gene function studies in the pitaya canker disease pathogen Neoscytalidium dimidiatum
- PMID: 36572711
- PMCID: PMC9792573
- DOI: 10.1038/s41598-022-27041-w
Identification of RT-qPCR reference genes suitable for gene function studies in the pitaya canker disease pathogen Neoscytalidium dimidiatum
Abstract
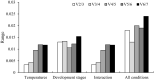
Neoscytalidium dimidiatum is the main causal agent of pitaya canker. Most studies of virulence and pathogenicity genes have measured expression levels using real-time quantitative polymerase chain reaction (RT-qPCR). Suitable reference genes are essential for ensuring that estimates of gene expression levels by RT-qPCR are accurate. However, no reference genes can be robustly applied across all contexts and species. No studies to date have evaluated the most effective reference genes for normalizing gene expression levels estimated by RT-qPCR in N. dimidiatum. In this study, RT-qPCR data for individual candidate reference genes were analyzed using four different methods: the delta Ct method and the geNorm, NormFinder, and BestKeeper algorithms. We evaluated the utility of eight candidate reference genes (18S rRNA, Actin (1), Actin (2), Actin, GAPDH (1), GAPDH (2), UBQ, and Tubulin) for normalizing expression levels estimated by RT-qPCR in N. dimidiatum at different developmental stages, at different temperatures, and during interaction with pitaya. All candidate reference genes were suitable for gene expression analysis except for Actin (2). Tubulin and Actin (1) were the most stably expressed reference genes under different temperatures. Actin (1) and Actin were the most stably expressed reference genes in N. dimidiatum at different developmental stages. Tubulin and UBQ were the most stably expressed reference genes during interaction with pitaya. Actin and 18s rRNA were the most stably expressed across all experimental conditions. Subsequently, Tubulin and UBQ were further investigated in analyses of pectinase expression during the pitaya-N. dimidiatum interaction. Our results provide insights that will aid future RT-qPCR studies of gene expression in N. dimidiatum.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Pan L, Zhang R, Lu F, Liu C, Fu J. Comparative proteomic analysis of the fungal pathogen Neoscytalidium dimidiatum infection in the pitaya. Hortic. Environ. Biotechnol. 2021;62:649–659. doi: 10.1007/s13580-021-00341-2. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

