A large language model for electronic health records
- PMID: 36572766
- PMCID: PMC9792464
- DOI: 10.1038/s41746-022-00742-2
A large language model for electronic health records
Abstract
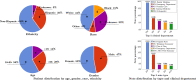
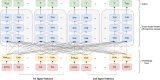
There is an increasing interest in developing artificial intelligence (AI) systems to process and interpret electronic health records (EHRs). Natural language processing (NLP) powered by pretrained language models is the key technology for medical AI systems utilizing clinical narratives. However, there are few clinical language models, the largest of which trained in the clinical domain is comparatively small at 110 million parameters (compared with billions of parameters in the general domain). It is not clear how large clinical language models with billions of parameters can help medical AI systems utilize unstructured EHRs. In this study, we develop from scratch a large clinical language model-GatorTron-using >90 billion words of text (including >82 billion words of de-identified clinical text) and systematically evaluate it on five clinical NLP tasks including clinical concept extraction, medical relation extraction, semantic textual similarity, natural language inference (NLI), and medical question answering (MQA). We examine how (1) scaling up the number of parameters and (2) scaling up the size of the training data could benefit these NLP tasks. GatorTron models scale up the clinical language model from 110 million to 8.9 billion parameters and improve five clinical NLP tasks (e.g., 9.6% and 9.5% improvement in accuracy for NLI and MQA), which can be applied to medical AI systems to improve healthcare delivery. The GatorTron models are publicly available at: https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/models/gatortron_og .
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals: 2008–2015. ONC Data Brief. https://www.healthit.gov/sites/default/files/briefs/2015_hospital_adopti... (2016).
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

