Unraveling the genetics underlying micronutrient signatures of diversity panel present in brown rice through genome-ionome linkages
- PMID: 36573652
- PMCID: PMC10952705
- DOI: 10.1111/tpj.16080
Unraveling the genetics underlying micronutrient signatures of diversity panel present in brown rice through genome-ionome linkages
Abstract
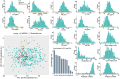
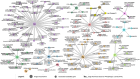
Rice (Oryza sativa) is an important staple crop to address the Hidden Hunger problem not only in Asia but also in Africa where rice is fast becoming an important source of calories. The brown rice (whole grain with bran) is known to be more nutritious due to elevated mineral composition. The genetics underlying brown rice ionome (sum total of such mineral composition) remains largely unexplored. Hence, we conducted a comprehensive study to dissect the genetic architecture of the brown rice ionome. We used genome-wide association studies, gene set analysis, and targeted association analysis for 12 micronutrients in the brown rice grains. A diverse panel of 300 resequenced indica accessions, with more than 1.02 million single nucleotide polymorphisms, was used. We identified 109 candidate genes with 5-20% phenotypic variation explained for the 12 micronutrients and identified epistatic interactions with multiple micronutrients. Pooling all candidate genes per micronutrient exhibited phenotypic variation explained values ranging from 11% to almost 40%. The key donor lines with larger concentrations for most of the micronutrients possessed superior alleles, which were absent in the breeding lines. Through gene regulatory networks we identified enriched functional pathways for central regulators that were detected as key candidate genes through genome-wide association studies. This study provided important insights on the ionome variations in rice, on the genetic basis of the genome-ionome relationships and on the molecular mechanisms underlying micronutrient signatures.
Keywords: GWAS; biofortification; epigenetics; gene regulatory network; micronutrients; systems-genetics.
© 2022 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
We declare no conflict of interest regarding this manuscript.
Figures





References
-
- Barrett, J.C. , Fry, B. , Maller, J. & Daly, M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. - PubMed
-
- Benjamini, Y. & Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, 57(1), 289–300 https://www.jstor.org/stable/2346101/
-
- Bollinedi, H. , Yadav, A.K. , Vinod, K.K. , Gopala, K.S. , Bhowmick, P.K. , Nagarajan, M. et al. (2020) Genome‐wide association study reveals novel marker‐trait associations (MTAs) governing the localization of Fe and Zn in the rice grain. Frontiers in Genetics, 11, 213. Available from: 10.3389/fgene.2020.00213 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

