Genomic Microevolution of Vibrio cholerae O1, Lake Tanganyika Basin, Africa
- PMID: 36573719
- PMCID: PMC9796204
- DOI: 10.3201/eid2901.220641
Genomic Microevolution of Vibrio cholerae O1, Lake Tanganyika Basin, Africa
Abstract
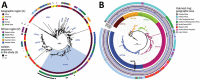
Africa's Lake Tanganyika basin is a cholera hotspot. During 2001-2020, Vibrio cholerae O1 isolates obtained from the Democratic Republic of the Congo side of the lake belonged to 2 of the 5 clades of the AFR10 sublineage. One clade became predominant after acquiring a parC mutation that decreased susceptibility to ciprofloxacin.
Keywords: Africa. Emerg Infect Dis. 2023 Jan [date cited]. https://doi.org/10.3201/eid2901.220641; African Great Lakes Region; Burundi; Democratic Republic of Congo; Gallandat K; Jeandron A; Kamwiziku G; Lake Tanganyika; Lake Tanganyika Basin; Njamkepo E; Rauzier J; Suggested citation for this article: Hounmanou YMG; Tanzania; Vibrio cholerae; Zambia; bacteria; cholera; et al. Genomic microevolution of Vibrio cholerae O1; genomics; neglected tropical diseases.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

