Global Gene Expression Analysis Reveals Complex Cuticle Organization of the Tribolium Compound Eye
- PMID: 36575057
- PMCID: PMC9866248
- DOI: 10.1093/gbe/evac181
Global Gene Expression Analysis Reveals Complex Cuticle Organization of the Tribolium Compound Eye
Abstract
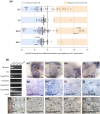
The red flour beetle Tribolium castaneum is a resource-rich model for genomic and developmental studies. To extend previous studies on Tribolium eye development, we produced transcriptomes for normal-eyed and eye-depleted heads of pupae and adults to identify differentially transcript-enriched (DE) genes in the visual system. Unexpectedly, cuticle-related genes were the largest functional class in the pupal compound eye DE gene population, indicating differential enrichment in three distinct cuticle components: clear lens facet cuticle, highly melanized cuticle of the ocular diaphragm, which surrounds the Tribolium compound eye for internal fortification, and newly identified facet margins of the tanned cuticle, possibly enhancing external fortification. Phylogenetic, linkage, and high-throughput gene knockdown data suggest that most cuticle proteins (CPs) expressed in the Tribolium compound eye stem from the deployment of ancient CP genes. Consistent with this, TcasCPR15, which we identified as the major lens CP gene in Tribolium, is a beetle-specific but pleiotropic paralog of the ancient CPR RR-2 CP gene family. The less abundant yet most likely even more lens-specific TcasCP63 is a member of a sprawling family of noncanonical CP genes, documenting a role of local gene family expansions in the emergence of the Tribolium compound eye CP repertoire. Comparisons with Drosophila and the mosquito Anopheles gambiae reveal a steady turnover of lens-enriched CP genes during insect evolution.
Keywords: Tribolium; cuticle; evodevo; eye; gene family evolution; lens proteins.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

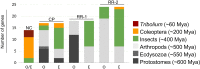





Similar articles
-
The beetle's structural protein CPCFC making elytra tough and rigid.Insect Sci. 2025 Jun;32(3):821-832. doi: 10.1111/1744-7917.13443. Epub 2024 Sep 5. Insect Sci. 2025. PMID: 39236247
-
Intraocular lens optic edge design for the prevention of posterior capsule opacification after cataract surgery.Cochrane Database Syst Rev. 2021 Aug 16;8(8):CD012516. doi: 10.1002/14651858.CD012516.pub2. Cochrane Database Syst Rev. 2021. PMID: 34398965 Free PMC article.
-
Lubricating drops for contact lens discomfort in adults.Cochrane Database Syst Rev. 2024 Sep 5;9(9):CD015751. doi: 10.1002/14651858.CD015751.pub2. Cochrane Database Syst Rev. 2024. PMID: 39234924 Free PMC article.
-
Manual small incision cataract surgery (MSICS) with posterior chamber intraocular lens versus phacoemulsification with posterior chamber intraocular lens for age-related cataract.Cochrane Database Syst Rev. 2013 Oct 10;2013(10):CD008813. doi: 10.1002/14651858.CD008813.pub2. Cochrane Database Syst Rev. 2013. PMID: 24114262 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
Cited by
-
Cataract induction in an arthropod reveals how lens crystallins contribute to the formation of biological glass.PLoS One. 2025 Jun 11;20(6):e0325229. doi: 10.1371/journal.pone.0325229. eCollection 2025. PLoS One. 2025. PMID: 40498792 Free PMC article.
-
The chromosome-level genome provides insights into the adaptive evolution of the visual system in Oratosquilla oratoria.BMC Biol. 2025 Feb 6;23(1):38. doi: 10.1186/s12915-025-02146-6. BMC Biol. 2025. PMID: 39915724 Free PMC article.
References
-
- Andersen SO, Chase AM, Willis JH. 1973. The amino-acid composition of cuticles from Tenebrio molitor with special reference to the action of juvenile hormone. Insect Biochem. 3:171–180.
-
- Arakane Y, et al. . 2005. The PLoS Genet. chitin synthase genes PLoS Genet. and PLoS Genet. are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. PLoS Genet. 14:453–463. - PubMed

