Single-cell transcriptomic analysis identifies murine heart molecular features at embryonic and neonatal stages
- PMID: 36575170
- PMCID: PMC9794824
- DOI: 10.1038/s41467-022-35691-7
Single-cell transcriptomic analysis identifies murine heart molecular features at embryonic and neonatal stages
Abstract
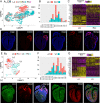
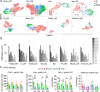
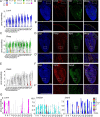
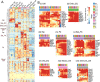
Heart development is a continuous process involving significant remodeling during embryogenesis and neonatal stages. To date, several groups have used single-cell sequencing to characterize the heart transcriptomes but failed to capture the progression of heart development at most stages. This has left gaps in understanding the contribution of each cell type across cardiac development. Here, we report the transcriptional profile of the murine heart from early embryogenesis to late neonatal stages. Through further analysis of this dataset, we identify several transcriptional features. We identify gene expression modules enriched at early embryonic and neonatal stages; multiple cell types in the left and right atriums are transcriptionally distinct at neonatal stages; many congenital heart defect-associated genes have cell type-specific expression; stage-unique ligand-receptor interactions are mostly between epicardial cells and other cell types at neonatal stages; and mutants of epicardium-expressed genes Wt1 and Tbx18 have different heart defects. Assessment of this dataset serves as an invaluable source of information for studies of heart development.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

