Drug sensitivity profiling of 3D tumor tissue cultures in the pediatric precision oncology program INFORM
- PMID: 36575299
- PMCID: PMC9794727
- DOI: 10.1038/s41698-022-00335-y
Drug sensitivity profiling of 3D tumor tissue cultures in the pediatric precision oncology program INFORM
Abstract
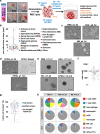
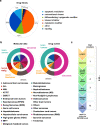
The international precision oncology program INFORM enrolls relapsed/refractory pediatric cancer patients for comprehensive molecular analysis. We report a two-year pilot study implementing ex vivo drug sensitivity profiling (DSP) using a library of 75-78 clinically relevant drugs. We included 132 viable tumor samples from 35 pediatric oncology centers in seven countries. DSP was conducted on multicellular fresh tumor tissue spheroid cultures in 384-well plates with an overall mean processing time of three weeks. In 89 cases (67%), sufficient viable tissue was received; 69 (78%) passed internal quality controls. The DSP results matched the identified molecular targets, including BRAF, ALK, MET, and TP53 status. Drug vulnerabilities were identified in 80% of cases lacking actionable (very) high-evidence molecular events, adding value to the molecular data. Striking parallels between clinical courses and the DSP results were observed in selected patients. Overall, DSP in clinical real-time is feasible in international multicenter precision oncology programs.
© 2022. The Author(s).
Conflict of interest statement
C.M.v.T. and I. Øra participated in the advisory boards of Novartis and Bayer. S.M.P. receives funding from Bayer, Pfizer, Eli-Lilly, Roche, Amgen, Astra Zeneca, PharmaMar, Sanofi and Servier in the context of an IMI-2-funded EU project entitled ITCC-P4 (
Figures






References
-
- van Tilburg CM, et al. The Pediatric Precision Oncology INFORM Registry: Clinical Outcome and Benefit for Patients with Very High-Evidence Targets. Cancer Discov. 2021;11:2764–2779. doi: 10.1158/2159-8290.CD-21-0094. - DOI - PMC - PubMed
-
- Newman S, et al. Genomes for kids: the scope of pathogenic mutations in pediatric cancer revealed by comprehensive DNA and RNA sequencing. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1631. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

