A comparison of marker-based estimators of inbreeding and inbreeding depression
- PMID: 36575379
- PMCID: PMC9793638
- DOI: 10.1186/s12711-022-00772-0
A comparison of marker-based estimators of inbreeding and inbreeding depression
Abstract
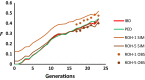
Background: The availability of genome-wide marker data allows estimation of inbreeding coefficients (F, the probability of identity-by-descent, IBD) and, in turn, estimation of the rate of inbreeding depression (ΔID). We investigated, by computer simulations, the accuracy of the most popular estimators of inbreeding based on molecular markers when computing F and ΔID in populations under random mating, equalization of parental contributions, and artificially selected populations. We assessed estimators described by Li and Horvitz (FLH1 and FLH2), VanRaden (FVR1 and FVR2), Yang and colleagues (FYA1 and FYA2), marker homozygosity (FHOM), runs of homozygosity (FROH) and estimates based on pedigree (FPED) in comparison with estimates obtained from IBD measures (FIBD).
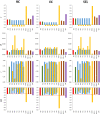
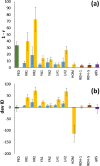
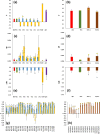
Results: If the allele frequencies of a base population taken as a reference for the computation of inbreeding are known, all estimators based on marker allele frequencies are highly correlated with FIBD and provide accurate estimates of the mean ΔID. If base population allele frequencies are unknown and current frequencies are used in the estimations, the largest correlation with FIBD is generally obtained by FLH1 and the best estimator of ΔID is FYA2. The estimators FVR2 and FLH2 have the poorest performance in most scenarios. The assumption that base population allele frequencies are equal to 0.5 results in very biased estimates of the average inbreeding coefficient but they are highly correlated with FIBD and give relatively good estimates of ΔID. Estimates obtained directly from marker homozygosity (FHOM) substantially overestimated ΔID. Estimates based on runs of homozygosity (FROH) provide accurate estimates of inbreeding and ΔID. Finally, estimates based on pedigree (FPED) show a lower correlation with FIBD than molecular estimators but provide rather accurate estimates of ΔID. An analysis of data from a pig population supports the main findings of the simulations.
Conclusions: When base population allele frequencies are known, all marker-allele frequency-based estimators of inbreeding coefficients generally show a high correlation with FIBD and provide good estimates of ΔID. When base population allele frequencies are unknown, FLH1 is the marker frequency-based estimator that is most correlated with FIBD, and FYA2 provides the most accurate estimates of ΔID. Estimates from FROH are also very precise in most scenarios. The estimators FVR2 and FLH2 have the poorest performances.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. 2. Cambridge: Cambridge University Press; 2010.
-
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sunderland: Sinauer Associates Inc.; 1998.
-
- Hedrick PW. Genetics of populations. Sudbury: Jones and Bartlett Publishers; 2010.
-
- Caballero A. Quantitative genetics. Cambridge: Cambridge University Press; 2020.
-
- Wright S. Coefficients of inbreeding and relationships. Am Nat. 1922;56:330–339. doi: 10.1086/279872. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

