Gut microbe Lactiplantibacillus plantarum undergoes different evolutionary trajectories between insects and mammals
- PMID: 36575413
- PMCID: PMC9795633
- DOI: 10.1186/s12915-022-01477-y
Gut microbe Lactiplantibacillus plantarum undergoes different evolutionary trajectories between insects and mammals
Abstract
Background: Animals form complex symbiotic associations with their gut microbes, whose evolution is determined by an intricate network of host and environmental factors. In many insects, such as Drosophila melanogaster, the microbiome is flexible, environmentally determined, and less diverse than in mammals. In contrast, mammals maintain complex multispecies consortia that are able to colonize and persist in the gastrointestinal tract. Understanding the evolutionary and ecological dynamics of gut microbes in different hosts is challenging. This requires disentangling the ecological factors of selection, determining the timescales over which evolution occurs, and elucidating the architecture of such evolutionary patterns.
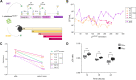
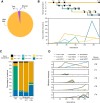
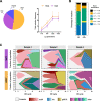
Results: We employ experimental evolution to track the pace of the evolution of a common gut commensal, Lactiplantibacillus plantarum, within invertebrate (Drosophila melanogaster) and vertebrate (Mus musculus) hosts and their respective diets. We show that in Drosophila, the nutritional environment dictates microbial evolution, while the host benefits L. plantarum growth only over short ecological timescales. By contrast, in a mammalian animal model, L. plantarum evolution results to be divergent between the host intestine and its diet, both phenotypically (i.e., host-evolved populations show higher adaptation to the host intestinal environment) and genomically. Here, both the emergence of hypermutators and the high persistence of mutated genes within the host's environment strongly differed from the low variation observed in the host's nutritional environment alone.
Conclusions: Our results demonstrate that L. plantarum evolution diverges between insects and mammals. While the symbiosis between Drosophila and L. plantarum is mainly determined by the host diet, in mammals, the host and its intrinsic factors play a critical role in selection and influence both the phenotypic and genomic evolution of its gut microbes, as well as the outcome of their symbiosis.
Keywords: Drosophila melanogaster; Experimental evolution; Gut microbiota evolution; Host–microbe symbiosis; Lactiplantibacillus plantarum; Mouse; Whole genome sequencing.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

