In silico analysis of peroxidase from Luffa acutangula
- PMID: 36575654
- PMCID: PMC9789927
- DOI: 10.1007/s13205-022-03432-8
In silico analysis of peroxidase from Luffa acutangula
Abstract
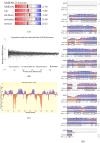
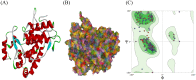
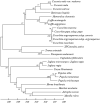
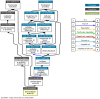
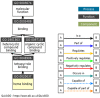
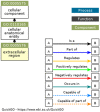
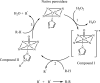
Peroxidases are oxidoreductase enzymes that widely gained attention as biocatalysts for their robust catalytic activity, specificity, and regioselective functionality for phenolic compounds. The study of molecular aspects of peroxidases is as crucial as that of the physicochemical aspects. A bioinformatics approach is utilized in this study to investigate the structural aspects and functions of luffa peroxidase (LPrx) from Luffa acutangula. The evolutionary relationship of LPrx with other class III peroxidases was studied by constructing a neighbour-joining phylogenetic tree. An analysis of the phylogenetic tree revealed that plant peroxidases share a common ancestor. The gene ontology term showed that LPrx had a molecular functionality of the oxidation-reduction process, heme binding and peroxidase-like activity, and the biological function of hydrogen peroxide scavenging activity. The enzyme-ligand interactions were studied from a catalytic point of view using the molecular docking technique. The molecular docking was carried out with LPrx as a receptor and guaiacol, m-cresol, p-cresol, catechol, quinol, pyrogallol, 2,4-dimethoxyphenol, gallic acid, aniline, and o-phenylenediamine as ligands. The results presented in the current communication will have a significant implication in proteomics, biochemistry, biotechnology, and the potential applications of peroxidases in the biotransformations of organic compounds.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03432-8.
Keywords: Enzyme functions; Gene ontology; Homology modelling; Molecular docking; Peroxidase; Phylogenesis.
© King Abdulaziz City for Science and Technology 2022, Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interest in any materials discussed in this article.
Figures



References
-
- Basumatary D, Yadav M, Nath P, Yadav HS. Catalytic biotransformations and inhibition study of peroxidase from Luffa aegyptiaca. Curr Organocatalysis. 2020 doi: 10.2174/2213337207666200211095038. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

