Retrograde signaling in plants: A critical review focusing on the GUN pathway and beyond
- PMID: 36575799
- PMCID: PMC9860301
- DOI: 10.1016/j.xplc.2022.100511
Retrograde signaling in plants: A critical review focusing on the GUN pathway and beyond
Abstract
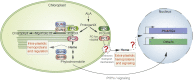
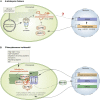
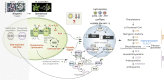
Plastids communicate their developmental and physiological status to the nucleus via retrograde signaling, allowing nuclear gene expression to be adjusted appropriately. Signaling during plastid biogenesis and responses of mature chloroplasts to environmental changes are designated "biogenic" and "operational" controls, respectively. A prominent example of the investigation of biogenic signaling is the screen for gun (genomes uncoupled) mutants. Although the first five gun mutants were identified 30 years ago, the functions of GUN proteins in retrograde signaling remain controversial, and that of GUN1 is hotly disputed. Here, we provide background information and critically discuss recently proposed concepts that address GUN-related signaling and some novel gun mutants. Moreover, considering heme as a candidate in retrograde signaling, we revisit the spatial organization of heme biosynthesis and export from plastids. Although this review focuses on GUN pathways, we also highlight recent progress in the identification and elucidation of chloroplast-derived signals that regulate the acclimation response in green algae and plants. Here, stress-induced accumulation of unfolded/misassembled chloroplast proteins evokes a chloroplast-specific unfolded protein response, which leads to changes in the expression levels of nucleus-encoded chaperones and proteases to restore plastid protein homeostasis. We also address the importance of chloroplast-derived signals for activation of flavonoid biosynthesis leading to production of anthocyanins during stress acclimation through sucrose non-fermenting 1-related protein kinase 1. Finally, a framework for identification and quantification of intercompartmental signaling cascades at the proteomic and metabolomic levels is provided, and we discuss future directions of dissection of organelle-nucleus communication.
Keywords: chloroplast; chloroplast unfolded protein response; genomes uncoupled; gun; retrograde signaling; subcellular metabolomes; subcellular proteomes.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Arrivault S., Guenther M., Florian A., Encke B., Feil R., Vosloh D., Lunn J.E., Sulpice R., Fernie A.R., Stitt M., et al. Dissecting the subcellular compartmentation of proteins and metabolites in arabidopsis leaves using non-aqueous fractionation. Mol. Cell. Proteomics. 2014;13:2246–2259. doi: 10.1074/mcp.M114.038190. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

