An ImageJ-based tool for three-dimensional registration between different types of microscopic images
- PMID: 36576380
- PMCID: PMC10107647
- DOI: 10.1111/dgd.12835
An ImageJ-based tool for three-dimensional registration between different types of microscopic images
Abstract
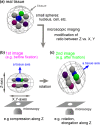
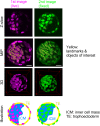
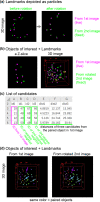
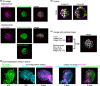
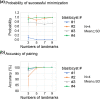
Three-dimensional (3D) registration (i.e., alignment) between two microscopic images is very helpful to study tissues that do not adhere to substrates, such as mouse embryos and organoids, which are often 3D rotated during imaging. However, there is no 3D registration tool easily accessible for experimental biologists. Here we developed an ImageJ-based tool which allows for 3D registration accompanied with both quantitative evaluation of the accuracy and reconstruction of 3D rotated images. In this tool, several landmarks are manually provided in two images to be aligned, and 3D rotation is computed so that the distances between the paired landmarks from the two images are minimized. By simultaneously providing multiple points (e.g., all nuclei in the regions of interest) other than the landmarks in the two images, the correspondence of each point between the two images, i.e., to which nucleus in one image a certain nucleus in another image corresponds, is quantitatively explored. Furthermore, 3D rotation is applied to one of the two images, resulting in reconstruction of 3D rotated images. We demonstrated that this tool successfully achieved 3D registration and reconstruction of images in mouse pre- and post-implantation embryos, where one image was obtained during live imaging and another image was obtained from fixed embryos after live imaging. This approach provides a versatile tool applicable for various tissues and species.
Keywords: ImageJ; image registration; mouse early embryo; three-dimensional image rotation.
© 2022 The Authors. Development, Growth & Differentiation published by John Wiley & Sons Australia, Ltd on behalf of Japanese Society of Developmental Biologists.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Automatic three-dimensional segmentation of mouse embryonic stem cell nuclei by utilising multiple channels of confocal fluorescence images.J Microsc. 2021 Jan;281(1):57-75. doi: 10.1111/jmi.12949. Epub 2020 Aug 8. J Microsc. 2021. PMID: 32720710
-
Registration of serial sections of mouse liver cell nuclei.J Microsc. 1998 Oct;192(Pt 1):37-53. doi: 10.1046/j.1365-2818.1998.00405.x. J Microsc. 1998. PMID: 9848269
-
Three-dimensional quantitative evaluation method of nonrigid registration algorithms for adaptive radiotherapy.Med Phys. 2010 Mar;37(3):1137-45. doi: 10.1118/1.3302916. Med Phys. 2010. PMID: 20384249
-
Multimodal 3D ultrasound and CT in image-guided spinal surgery: public database and new registration algorithms.Int J Comput Assist Radiol Surg. 2021 Apr;16(4):555-565. doi: 10.1007/s11548-021-02323-2. Epub 2021 Mar 8. Int J Comput Assist Radiol Surg. 2021. PMID: 33683544 Review.
-
Rapid high resolution three dimensional reconstruction of embryos with episcopic fluorescence image capture.Birth Defects Res C Embryo Today. 2004 Sep;72(3):213-23. doi: 10.1002/bdrc.20023. Birth Defects Res C Embryo Today. 2004. PMID: 15495188 Review.
References
-
- Fernandez, R. , & Moisy, C. (2021). Fijiyama: A registration tool for 3D multimodal time‐lapse imaging. Bioinformatics, 37, 1482–1484. - PubMed
MeSH terms
Grants and funding
- Japan Ministry of Education, Culture, Sports, Science and Technology Grant-in-Aid for Scientific Research on Innovative Areas "Cross-talk between moving cells and microenvironment as a basis of emerging order"
- JPMJCR1654/Japan Science and Technology Agency, Core Research for Evolutional Science and Technology (JST-CREST) grant
- 17K15131/Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Young Scientists (B)
LinkOut - more resources
Full Text Sources

