Deep learning-driven insights into super protein complexes for outer membrane protein biogenesis in bacteria
- PMID: 36576775
- PMCID: PMC9797188
- DOI: 10.7554/eLife.82885
Deep learning-driven insights into super protein complexes for outer membrane protein biogenesis in bacteria
Abstract
To reach their final destinations, outer membrane proteins (OMPs) of gram-negative bacteria undertake an eventful journey beginning in the cytosol. Multiple molecular machines, chaperones, proteases, and other enzymes facilitate the translocation and assembly of OMPs. These helpers usually associate, often transiently, forming large protein assemblies. They are not well understood due to experimental challenges in capturing and characterizing protein-protein interactions (PPIs), especially transient ones. Using AF2Complex, we introduce a high-throughput, deep learning pipeline to identify PPIs within the Escherichia coli cell envelope and apply it to several proteins from an OMP biogenesis pathway. Among the top confident hits obtained from screening ~1500 envelope proteins, we find not only expected interactions but also unexpected ones with profound implications. Subsequently, we predict atomic structures for these protein complexes. These structures, typically of high confidence, explain experimental observations and lead to mechanistic hypotheses for how a chaperone assists a nascent, precursor OMP emerging from a translocon, how another chaperone prevents it from aggregating and docks to a β-barrel assembly port, and how a protease performs quality control. This work presents a general strategy for investigating biological pathways by using structural insights gained from deep learning-based predictions.
Keywords: E. coli; computational biology; deep learning; molecular biophysics; outer membrane protein biogenesis; protein complex structure prediction; protein-protein interaction; structural biology; systems biology; translocon; virtual screening.
Plain language summary
All living cells are contained within a fatty cell membrane that allows water and only certain other molecules to pass through with ease. Bacteria only consist of a single cell, making their membrane the only interface with the surrounding environment. Gram-negative bacteria – which include Escherichia coli, a bacterium found in the gut of all humans – have an extra layer of protection, the ‘outer membrane’. Proteins in this membrane are called ‘outer membrane proteins’ or OMPs and allow nutrients to enter the cell. But OMPs, which are made inside the cell, need to be transported to the outer membrane and folded correctly before they can perform their role. This multistep process, which involves interactions between many different proteins, is not fully understood. The journey of an OMP from the center of the cell where it is made to the outer membrane is complicated. First, the OMP needs to pass through the cell’s inner membrane. To do this, it must interact with ‘channel proteins’ in the inner membrane that feed the OMP into the space between the two membranes, known as the bacterial envelope. This step requires the OMP to be unfolded. Once in the bacterial envelope the OMP interacts with proteins that help it fold correctly and integrate into the outer membrane. The interactions between proteins in the bacterial envelope are short-lived, making them difficult to study using lab-based experiments. An alternative approach is predicting a protein’s structure from its amino acid sequence which is a difficult computational problem to solve. However, in 2020 developers behind the AlphaFold2, a deep learning program, were able to use a set of equations organized in a ‘neural network’ that can ‘learn’ from a library of known protein structures to predict unknown structures with high accuracy. Gao et al. used AF2Complex, a tool based AlphaFold2, tailored to predicting interactions between proteins, to investigate what interactions OMPs could be involved with on their way to the outer membrane. With the help of a supercomputer at the Oakridge National Laboratory, Gao et al. screened nearly 1,500 E. coli proteins within the bacterial envelope to see how they might interact with OMPs. The screen identified previously unknown interactions between proteins that suggest that the formation of the bacterial outer membrane and the integration of proteins into it involve protein complexes and molecular mechanisms that have not yet been characterized. Additionally, the screen also identified interactions that had been previously described, confirming that the deep learning approach can correctly capture real interactions. Overall, Gao et al.’s work inspires new hypotheses about the mechanisms through which OMPs are transported to the outer membrane, although further work will be needed to confirm the roles of protein interactions predicted by the computational model experimentally. Furthermore, the ability to design experiments based on computational predictions is exciting. If confirmed, the new protein interactions could help scientists better understand OMP transport, which is essential for bacterial biology. In the future, this could lead to the discovery of new targets for antibiotic drugs.
© 2022, Gao et al.
Conflict of interest statement
MG, DN, JS No competing interests declared
Figures

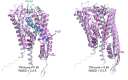

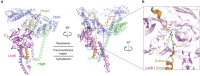

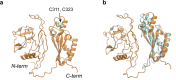




References
-
- Babu M, Bundalovic-Torma C, Calmettes C, Phanse S, Zhang Q, Jiang Y, Minic Z, Kim S, Mehla J, Gagarinova A, Rodionova I, Kumar A, Guo H, Kagan O, Pogoutse O, Aoki H, Deineko V, Caufield JH, Holtzapple E, Zhang Z, Vastermark A, Pandya Y, Lai CC-L, El Bakkouri M, Hooda Y, Shah M, Burnside D, Hooshyar M, Vlasblom J, Rajagopala SV, Golshani A, Wuchty S, F Greenblatt J, Saier M, Uetz P, F Moraes T, Parkinson J, Emili A. Global landscape of cell envelope protein complexes in Escherichia coli. Nature Biotechnology. 2018;36:103–112. doi: 10.1038/nbt.4024. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

