Identification of COVID-19 spread mechanisms based on first-wave data, simulation models, and evolutionary algorithms
- PMID: 36576938
- PMCID: PMC9797101
- DOI: 10.1371/journal.pone.0279427
Identification of COVID-19 spread mechanisms based on first-wave data, simulation models, and evolutionary algorithms
Abstract
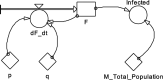
Background: The COVID-19 epidemic has shown that efficient prediction models are required, and the well-known SI, SIR, and SEIR models are not always capable of capturing the real dynamics. Modified models with novel structures could help identify unknown mechanisms of COVID-19 spread.
Objective: Our objective is to provide additional insights into the COVID-19 spread mechanisms based on different models' parameterization which was performed using evolutionary algorithms and the first-wave data.
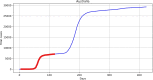
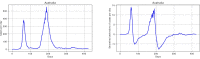
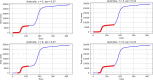
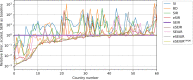
Methods: Data from the Our World in Data COVID-19 database was analysed, and several models-SI, SIR, SEIR, SEIUR, and Bass diffusion-and their variations were considered for the first wave of the COVID-19 pandemic. The models' parameters were tuned with differential evolution optimization method L-SHADE to find the best fit. The algorithm for the automatic identification of the first wave was developed, and the differential evolution was applied to model parameterization. The reproduction rates (R0) for the first wave were calculated for 61 countries based on the best fits.
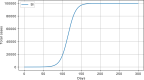
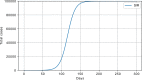
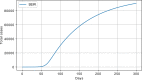
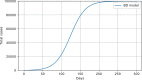
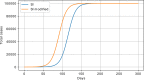
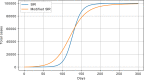
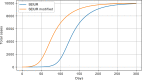
Results: The performed experiments showed that the Bass diffusion model-based modification could be superior compared to SI, SIR, SEIR and SEIUR due to the component responsible for spread from an external factor, which is not directly dependent on contact with infected individuals. The developed modified models containing this component were shown to perform better when fitting to the first-wave cumulative infections curve. In particular, the modified SEIR model was better fitted to the real-world data than the classical SEIR in 43 cases out of 61, based on Mann-Whitney U tests; the Bass diffusion model was better than SI for 57 countries. This showed the limitation of the classical models and indicated ways to improve them.
Conclusions: By using the modified models, the mechanism of infection spread, which is not directly dependent on contacts, was identified, which significantly influences the dynamics of the spread of COVID-19.
Copyright: © 2022 Stanovov et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Hethcote H. The mathematics of infectious diseases. SIAM Rev 2000;42: 599–653. 10.1137/S0036144500371907 - DOI
-
- Kermack WO, McKendrick AG. A contribution to the mathematical theory of epidemics. Proceedings Royal Soc. London (A); 1927;115:700–721. 10.1098/rspa.1927.0118 - DOI
-
- Keeling MJ, Rohan P. Modelling infectious diseases. Princeton University Press; 2008. ISBN 9780691116174. 10.2307/j.ctvcm4gk0 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

