Translational Pharmacokinetic/Pharmacodynamic Model for mRNA-3927, an Investigational Therapeutic for the Treatment of Propionic Acidemia
- PMID: 36577040
- PMCID: PMC10066765
- DOI: 10.1089/nat.2022.0036
Translational Pharmacokinetic/Pharmacodynamic Model for mRNA-3927, an Investigational Therapeutic for the Treatment of Propionic Acidemia
Abstract
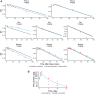
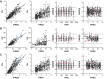
Propionic acidemia (PA) is an ultrarare disorder caused by deficiency of the mitochondrial enzyme, propionyl-CoA carboxylase (PCC), composed of PCCA and PCCB subunits. An enzyme replacement therapy is being developed using dual messenger RNA (mRNA) therapy composed of lipid nanoparticles (LNPs) encapsulating mRNAs encoding PCCA and PCCB subunits of the PCC enzyme. We herein report on development of a translational semimechanistic pharmacokinetic (PK) and PK/pharmacodynamic (PD) model to quantify the relationship between the mRNA components of mRNA-3927 (an LNP encapsulating PCCA and PCCB mRNAs) and dose levels; PCCA/B mRNA PK and PD responses were assessed as circulating levels of primary disease markers 2-methyl citrate, 3-hydroxypropionate, and propionyl carnitine normalized to acetyl carnitine (C3/C2 ratio) to inform the first-in-human dose range and regimen selection. The translational PK/PD model was developed using preclinical data available in mice with PA, Sprague Dawley rats, and cynomolgus monkeys at dose levels ranging from 0.2 to 9 mg/kg. PCCA/B mRNA PK in mice, rats, and monkeys was adequately described using allometric scaling of volume and clearance parameters. The interspecies preclinical model was scaled allometrically to humans to predict the dose-response relationship in adult and pediatric patients with PA to guide selection of dose range and regimen for the Phase 1 clinical trial (ClinicalTrials.gov Identifier NCT04159103).
Keywords: lipid nanoparticles; mRNA therapy; propionic acidemia; propionyl-CoA carboxylase.
Conflict of interest statement
H.A., M.L., M.L., and J.S. are employees of Moderna, Inc., and hold stock/options in the company. V.I. is an employee of Pumas AI.
Figures


References
-
- Zwickler T, Riderer A, Haege G, Hoffmann GF, Kolker S and Burgard P. (2014). Usefulness of biochemical parameters in decision-making on the start of emergency treatment in patients with propionic acidemia. J Inherit Metab Dis 37:31–37. - PubMed
-
- Ugarte M, Pérez-Cerdá C, Rodríguez-Pombo P, Desviat LR, Pérez B, Richard E, Muro S, Campeau E, Ohura T and Gravel RA. (1999). Overview of mutations in the PCCA and PCCB genes causing propionic acidemia. Human Mutat 14:275–282. - PubMed
-
- Jiang H, Rao KS, Yee VC and Kraus JP. (2005). Characterization of four variant forms of human propionyl-CoA carboxylase expressed in Escherichia coli. J Biol Chem 280:27719–27727. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
