Staged suppression of microglial autophagy facilitates regeneration in CNS demyelination by enhancing the production of linoleic acid
- PMID: 36577069
- PMCID: PMC9910603
- DOI: 10.1073/pnas.2209990120
Staged suppression of microglial autophagy facilitates regeneration in CNS demyelination by enhancing the production of linoleic acid
Abstract
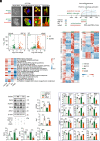
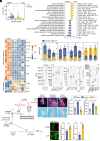
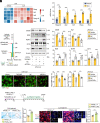
Microglia play a critical role in the clearance of myelin debris, thereby ensuring functional recovery from neural injury. Here, using mouse model of demyelination following two-point LPC injection, we show that the microglial autophagic-lysosomal pathway becomes overactivated in response to severe demyelination, leading to lipid droplet accumulation and a dysfunctional and pro-inflammatory microglial state, and finally failed myelin debris clearance and spatial learning deficits. Data from genetic approaches and pharmacological modulations, via microglial Atg5 deficient mice and intraventricular BAF A1 administration, respectively, demonstrate that staged suppression of excessive autophagic-lysosomal activation in microglia, but not sustained inhibition, results in better myelin debris degradation and exerts protective effects against demyelination. Combined multi-omics results in vitro further showed that enhanced lipid metabolism, especially the activation of the linoleic acid pathway, underlies this protective effect. Supplementation with conjugated linoleic acid (CLA), both in vivo and in vitro, could mimic these effects, including attenuating inflammation and restoring microglial pro-regenerative properties, finally resulting in better recovery from demyelination injuries and improved spatial learning function, by activating the peroxisome proliferator-activated receptor (PPAR-γ) pathway. Therefore, we propose that pharmacological inhibition targeting microglial autophagic-lysosomal overactivation or supplementation with CLA could represent a potential therapeutic strategy in demyelinated disorders.
Keywords: autophagic–lysosomal pathway; conjugated linoleic acid; demyelination; lipid metabolism; microglia.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Haste makes waste: staged suppression of autophagic-lysosomal pathway in microglia promotes the efficient clearance of myelin debris.Autophagy. 2023 Jun;19(6):1896-1898. doi: 10.1080/15548627.2023.2186097. Epub 2023 Mar 5. Autophagy. 2023. PMID: 36871202 Free PMC article.
-
Deficiency of microglial Hv1 channel is associated with activation of autophagic pathway and ROS production in LPC-induced demyelination mouse model.J Neuroinflammation. 2020 Nov 6;17(1):333. doi: 10.1186/s12974-020-02020-y. J Neuroinflammation. 2020. PMID: 33158440 Free PMC article.
-
Transforming Growth Factor β1 Protects Against Ischemic Demyelination via Regulating Microglial Lipid Metabolism Pathway.Stroke. 2025 Jun;56(6):1554-1568. doi: 10.1161/STROKEAHA.124.048206. Epub 2025 Mar 31. Stroke. 2025. PMID: 40160039
-
The roles of microglia and astrocytes in phagocytosis and myelination: Insights from the cuprizone model of multiple sclerosis.Glia. 2022 Jul;70(7):1215-1250. doi: 10.1002/glia.24148. Epub 2022 Feb 2. Glia. 2022. PMID: 35107839 Free PMC article. Review.
-
Neuronal injury in chronic CNS inflammation.Best Pract Res Clin Anaesthesiol. 2010 Dec;24(4):551-62. doi: 10.1016/j.bpa.2010.11.001. Epub 2010 Nov 29. Best Pract Res Clin Anaesthesiol. 2010. PMID: 21619866 Review.
Cited by
-
Anti-neuroinflammatory effects of conjugated linoleic acid isomers, c9,t11 and t10,c12, on activated BV-2 microglial cells.Front Cell Neurosci. 2024 Sep 27;18:1442786. doi: 10.3389/fncel.2024.1442786. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39398647 Free PMC article.
-
Nervonic Acid Synthesis Substrates as Essential Components in Profiled Lipid Supplementation for More Effective Central Nervous System Regeneration.Int J Mol Sci. 2024 Mar 28;25(7):3792. doi: 10.3390/ijms25073792. Int J Mol Sci. 2024. PMID: 38612605 Free PMC article. Review.
-
Ido2 Deficiency Exacerbates Motor Impairment and Reduces Aryl Hydrocarbon Receptor Activity through Decreased Kynurenine in a Chronic Demyelinating Mouse Model.Mol Neurobiol. 2025 Jan;62(1):109-122. doi: 10.1007/s12035-024-04263-9. Epub 2024 Jun 3. Mol Neurobiol. 2025. PMID: 38829509
-
Lysosomal acidification dysfunction in microglia: an emerging pathogenic mechanism of neuroinflammation and neurodegeneration.J Neuroinflammation. 2023 Aug 5;20(1):185. doi: 10.1186/s12974-023-02866-y. J Neuroinflammation. 2023. PMID: 37543564 Free PMC article. Review.
-
Astragaloside's Role in Microglial Neuroinflammation: A Multi-Method Study Combining Experimental and Computational Analyses.Neurochem Res. 2025 Jun 23;50(4):205. doi: 10.1007/s11064-025-04459-y. Neurochem Res. 2025. PMID: 40549030
References
-
- Ballabio A., Bonifacino J. S., Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118 (2019). - PubMed
-
- Berglund R., et al. , Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci. Immunol. 5, eabb5077 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

